A Role for the RNA Polymerase Gene Specificity Factor σ54 in the Uniform Colony Growth of Uropathogenic Escherichia coli
- PMID: 35357162
- PMCID: PMC9017345
- DOI: 10.1128/jb.00031-22
A Role for the RNA Polymerase Gene Specificity Factor σ54 in the Uniform Colony Growth of Uropathogenic Escherichia coli
Abstract
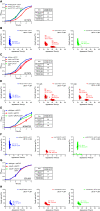
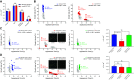
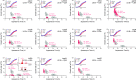
The canonical function of a bacterial sigma (σ) factor is to determine the gene specificity of the RNA polymerase (RNAP). In several diverse bacterial species, the σ54 factor uniquely confers distinct functional and regulatory properties on the RNAP. A hallmark feature of the σ54-RNAP is the obligatory requirement for an activator ATPase to allow transcription initiation. Different activator ATPases couple diverse environmental cues to the σ54-RNAP to mediate adaptive changes in gene expression. Hence, the genes that rely upon σ54 for their transcription have a wide range of different functions suggesting that the repertoire of functions performed by genes, directly or indirectly affected by σ54, is not yet exhaustive. By comparing the growth patterns of prototypical enteropathogenic, uropathogenic, and nonpathogenic Escherichia coli strains devoid of σ54, we uncovered that the absence of σ54 results in two differently sized colonies that appear at different times specifically in the uropathogenic E. coli (UPEC) strain. Notably, UPEC bacteria devoid of individual activator ATPases of the σ54-RNAP do not phenocopy the σ54 mutant strain. Thus, it seems that σ54's role as a determinant of uniform colony appearance in UPEC bacteria represents a putative non-canonical function of σ54 in regulating genetic information flow. IMPORTANCE RNA synthesis is the first step of gene expression. The multisubunit RNA polymerase (RNAP) is the central enzyme responsible for RNA synthesis in bacteria. The dissociable sigma (σ) factor subunit directs the RNAP to different sets of genes to allow their expression in response to various cellular needs. Of the seven σ factors in Escherichia coli and related bacteria, σ54 exists in a class of its own. This study has uncovered that σ54 is a determinant of the uniform growth of uropathogenic E. coli on solid media. This finding suggests a role for this σ54 in gene regulation that extends beyond its known function as an RNAP gene specificity factor.
Keywords: Escherichia coli; RNA polymerase; UPEC; microcolonies; sigma 54; sigma factors; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
An Amino Acid Substitution in RNA Polymerase That Inhibits the Utilization of an Alternative Sigma Factor.J Bacteriol. 2017 Jun 27;199(14):e00277-17. doi: 10.1128/JB.00277-17. Print 2017 Jul 15. J Bacteriol. 2017. PMID: 28507241 Free PMC article.
-
Association of ω with the C-Terminal Region of the β' Subunit Is Essential for Assembly of RNA Polymerase in Mycobacterium tuberculosis.J Bacteriol. 2018 May 24;200(12):e00159-18. doi: 10.1128/JB.00159-18. Print 2018 Jun 15. J Bacteriol. 2018. PMID: 29632095 Free PMC article.
-
Protein-protein interactions between sigma(70) region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for "off-DNA" and "on-DNA" interactions.J Mol Biol. 2010 Aug 6;401(1):13-32. doi: 10.1016/j.jmb.2010.05.052. Epub 2010 Jun 2. J Mol Biol. 2010. PMID: 20595001 Free PMC article.
-
The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences.FEMS Microbiol Lett. 2000 May 1;186(1):1-9. doi: 10.1111/j.1574-6968.2000.tb09074.x. FEMS Microbiol Lett. 2000. PMID: 10779705 Review.
-
Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity.FEMS Microbiol Rev. 2010 Sep;34(5):646-57. doi: 10.1111/j.1574-6976.2010.00223.x. Epub 2010 Apr 14. FEMS Microbiol Rev. 2010. PMID: 20491934 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

