Spatial transcriptomic profiles of mouse uterine microenvironments at pregnancy day 7.5†
- PMID: 35357464
- PMCID: PMC9382390
- DOI: 10.1093/biolre/ioac061
Spatial transcriptomic profiles of mouse uterine microenvironments at pregnancy day 7.5†
Abstract
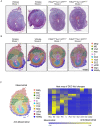
Uterine dysfunctions lead to fertility disorders and pregnancy complications. Normal uterine functions at pregnancy depend on crosstalk among multiple cell types in uterine microenvironments. Here, we performed the spatial transcriptomics and single-cell RNA-seq assays to determine local gene expression profiles at the embryo implantation site of the mouse uterus on pregnancy day 7.5 (D7.5). The spatial transcriptomic annotation identified 11 domains of distinct gene signatures, including a mesometrial myometrium, an anti-mesometrial myometrium, a mesometrial decidua enriched with natural killer cells, a vascular sinus zone for maternal vessel remodeling, a fetal-maternal interface, a primary decidual zone, a transition decidual zone, a secondary decidual zone, undifferentiated stroma, uterine glands, and the embryo. The scRNA-Seq identified 12 types of cells in the D7.5 uterus including three types of stromal fibroblasts with differentiated and undifferentiated markers, one cluster of epithelium including luminal and glandular epithelium, mesothelium, endothelia, pericytes, myelomonocytic cell, natural killer cells, and lymphocyte B. These single-cell RNA signatures were then utilized to deconvolute the cell-type compositions of each individual uterine microenvironment. Functional annotation assays on spatial transcriptomic data revealed uterine microenvironments with distinguished metabolic preferences, immune responses, and various cellular behaviors that are regulated by region-specific endocrine and paracrine signals. Global interactome among regions is also projected based on the spatial transcriptomic data. This study provides high-resolution transcriptome profiles with locality information at the embryo implantation site to facilitate further investigations on molecular mechanisms for normal pregnancy progression.
Keywords: decidua; microenvironment; postimplantation; scRNA-Seq; spatial transcriptomics; uterus.
Published by Oxford University Press on behalf of Society for the Study of Reproduction 2022.
Figures






References
-
- Farquhar CM, Bhattacharya S, Repping S, Mastenbroek S, Kamath MS, Marjoribanks J, Boivin J. Female subfertility. Nat Rev Dis Primers 2019; 5:7. - PubMed
-
- Law A, McCoy M, Lynen R, Curkendall SM, Gatwood J, Juneau PL, Landsman-Blumberg P. The prevalence of complications and healthcare costs during pregnancy. J Med Econ 2015; 18:533–541. - PubMed
-
- Tamaya T, Fujimoto J, Arabori K, Wada K, Kato Y, Okada H. The biologic role of sex steroid receptors in the decidualization of human endometrium. Asia Oceania J Obstet Gynaecol 1985; 11:573–578. - PubMed
-
- Dunk C, Kwan M, Hazan A, Walker S, Wright JK, Harris LK, Jones RL, Keating S, Kingdom JCP, Whittle W, Maxwell C and Lye SJ. Failure of decidualization and maternal immune tolerance underlies uterovascular resistance in intra uterine growth restriction. Front Endocrinol (Lausanne) 2019; 10:160. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

