Alkaline nucleoplasm facilitates contractile gene expression in the mammalian heart
- PMID: 35357563
- PMCID: PMC8971196
- DOI: 10.1007/s00395-022-00924-9
Alkaline nucleoplasm facilitates contractile gene expression in the mammalian heart
Abstract
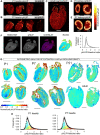
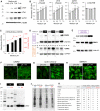
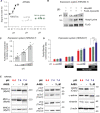
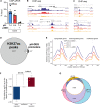
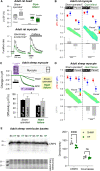
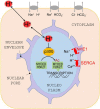
Cardiac contractile strength is recognised as being highly pH-sensitive, but less is known about the influence of pH on cardiac gene expression, which may become relevant in response to changes in myocardial metabolism or vascularization during development or disease. We sought evidence for pH-responsive cardiac genes, and a physiological context for this form of transcriptional regulation. pHLIP, a peptide-based reporter of acidity, revealed a non-uniform pH landscape in early-postnatal myocardium, dissipating in later life. pH-responsive differentially expressed genes (pH-DEGs) were identified by transcriptomics of neonatal cardiomyocytes cultured over a range of pH. Enrichment analysis indicated "striated muscle contraction" as a pH-responsive biological process. Label-free proteomics verified fifty-four pH-responsive gene-products, including contractile elements and the adaptor protein CRIP2. Using transcriptional assays, acidity was found to reduce p300/CBP acetylase activity and, its a functional readout, inhibit myocardin, a co-activator of cardiac gene expression. In cultured myocytes, acid-inhibition of p300/CBP reduced H3K27 acetylation, as demonstrated by chromatin immunoprecipitation. H3K27ac levels were more strongly reduced at promoters of acid-downregulated DEGs, implicating an epigenetic mechanism of pH-sensitive gene expression. By tandem cytoplasmic/nuclear pH imaging, the cardiac nucleus was found to exercise a degree of control over its pH through Na+/H+ exchangers at the nuclear envelope. Thus, we describe how extracellular pH signals gain access to the nucleus and regulate the expression of a subset of cardiac genes, notably those coding for contractile proteins and CRIP2. Acting as a proxy of a well-perfused myocardium, alkaline conditions are permissive for expressing genes related to the contractile apparatus.
Keywords: Acidity; CRIP2; Cardiomyocyte; Contraction; Nucleus.
© 2022. The Author(s).
Conflict of interest statement
OAA and YKR are founders of pHLIP, Inc. They have shares in the company, but the company did not fund any part of the work. TAM and NTC are shareholders in and consultants for Dark Blue Therapeutics Ltd.
Figures



References
-
- Baartscheer A, Schumacher CA, van Borren MM, Belterman CN, Coronel R, Fiolet JW. Increased Na+/H+-exchange activity is the cause of increased [Na+]i and underlies disturbed calcium handling in the rabbit pressure and volume overload heart failure model. Cardiovasc Res. 2003;57:1015–1024. doi: 10.1016/S0008-6363(02)00809-X. - DOI - PubMed
-
- Bkaily G, Nader M, Avedanian L, Jacques D, Perrault C, Abdel-Samad D, D'Orleans-Juste P, Gobeil F, Hazzouri KM. Immunofluorescence revealed the presence of NHE-1 in the nuclear membranes of rat cardiomyocytes and isolated nuclei of human, rabbit, and rat aortic and liver tissues. Can J Physiol Pharmacol. 2004;82:805–811. doi: 10.1139/y04-119. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

