Polyglutamylation: biology and analysis
- PMID: 35357568
- PMCID: PMC9117372
- DOI: 10.1007/s00726-022-03146-4
Polyglutamylation: biology and analysis
Abstract
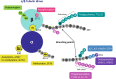
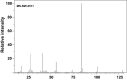
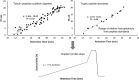
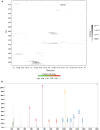
Polyglutamylation is a posttranslational modification (PTM) that adds several glutamates on glutamate residues in the form of conjugated peptide chains by a family of enzymes known as polyglutamylases. Polyglutamylation is well documented in microtubules. Polyglutamylated microtubules consist of different α- and β-tubulin subunits with varied number of added glutamate residues. Kinetic control and catalytic rates of tubulin modification by polyglutamylases influence the polyglutamylation pattern of functional microtubules. The recent studies uncovered catalytic mechanisms of the glutamylation enzymes family, particularly tubulin tyrosine ligase-like (TTLL). Variable length polyglutamylation of primary sequence glutamyl residues have been mapped with a multitude of protein chemistry and proteomics approaches. Although polyglutamylation was initially considered a tubulin-specific modification, the recent studies have uncovered a calmodulin-dependent glutamylase, SidJ. Nano-electrospray ionization (ESI) proteomic approaches have identified quantifiable polyglutamylated sites in specific substrates. Indeed, conjugated glutamylated peptides were used in nano-liquid chromatography gradient delivery due to their relative hydrophobicity for their tandem mass spectrometry (MS/MS) characterization. The recent polyglutamylation characterization has revealed three major sites: E445 in α-tubulin, E435 in β-tubulin, and E860 in SdeA. In this review, we have summarized the progress made using proteomic approaches for large-scale detection of polyglutamylated peptides, including biology and analysis.
Keywords: Mass spectrometry; NanoESI; Polyglutamylation; Protein chemistry; Proteomics; Tubulin.
© 2022. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures
Similar articles
-
Mass Spectrometry Reveals Novel Features of Tubulin Polyglutamylation in the Flagellum of Trypanosoma brucei.J Proteome Res. 2025 Aug 1;24(8):3979-3989. doi: 10.1021/acs.jproteome.5c00107. Epub 2025 Jul 9. J Proteome Res. 2025. PMID: 40634267
-
Mapping of polyglutamylation in tubulins using nanoLC-ESI-MS/MS.Anal Biochem. 2021 Jan 1;612:113761. doi: 10.1016/j.ab.2020.113761. Epub 2020 Jun 2. Anal Biochem. 2021. PMID: 32502490
-
Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on beta-tubulin through a random sequential pathway.Biochemistry. 2009 Feb 10;48(5):1084-93. doi: 10.1021/bi802047y. Biochemistry. 2009. PMID: 19152315
-
Potential role of tubulin tyrosine ligase-like enzymes in tumorigenesis and cancer cell resistance.Cancer Lett. 2014 Aug 1;350(1-2):1-4. doi: 10.1016/j.canlet.2014.04.022. Epub 2014 May 6. Cancer Lett. 2014. PMID: 24814394 Review.
-
Polyglutamylation: a fine-regulator of protein function? 'Protein Modifications: beyond the usual suspects' review series.EMBO Rep. 2008 Jul;9(7):636-41. doi: 10.1038/embor.2008.114. Epub 2008 Jun 20. EMBO Rep. 2008. PMID: 18566597 Free PMC article. Review.
Cited by
-
AGBL4 promotes malignant progression of glioblastoma via modulation of MMP-1 and inflammatory pathways.Front Immunol. 2024 Jun 28;15:1420182. doi: 10.3389/fimmu.2024.1420182. eCollection 2024. Front Immunol. 2024. PMID: 39007144 Free PMC article.
-
Editorial.Amino Acids. 2022 Apr;54(4):481-484. doi: 10.1007/s00726-022-03164-2. Amino Acids. 2022. PMID: 35486192 Free PMC article. No abstract available.
-
Barcoding Microtubules: Encoding Information onto Macromolecules by Photobleaching.Nano Lett. 2025 Apr 2;25(13):5283-5290. doi: 10.1021/acs.nanolett.5c00105. Epub 2025 Mar 21. Nano Lett. 2025. PMID: 40117582 Free PMC article.
-
Proximity Mapping of CCP6 Reveals Its Association with Centrosome Organization and Cilium Assembly.Int J Mol Sci. 2023 Jan 9;24(2):1273. doi: 10.3390/ijms24021273. Int J Mol Sci. 2023. PMID: 36674791 Free PMC article.
-
Netrin-1 stimulated axon growth requires the polyglutamylase TTLL1.Front Neurosci. 2024 Oct 14;18:1436312. doi: 10.3389/fnins.2024.1436312. eCollection 2024. Front Neurosci. 2024. PMID: 39469034 Free PMC article.
References
-
- Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc Natl Acad Sci USA. 1991;88(11):4685–4689. doi: 10.1073/pnas.88.11.4685. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

