Folliculin promotes substrate-selective mTORC1 activity by activating RagC to recruit TFE3
- PMID: 35358174
- PMCID: PMC9004751
- DOI: 10.1371/journal.pbio.3001594
Folliculin promotes substrate-selective mTORC1 activity by activating RagC to recruit TFE3
Abstract
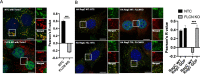
Mechanistic target of rapamycin complex I (mTORC1) is central to cellular metabolic regulation. mTORC1 phosphorylates a myriad of substrates, but how different substrate specificity is conferred on mTORC1 by different conditions remains poorly defined. Here, we show how loss of the mTORC1 regulator folliculin (FLCN) renders mTORC1 specifically incompetent to phosphorylate TFE3, a master regulator of lysosome biogenesis, without affecting phosphorylation of other canonical mTORC1 substrates, such as S6 kinase. FLCN is a GTPase-activating protein (GAP) for RagC, a component of the mTORC1 amino acid (AA) sensing pathway, and we show that active RagC is necessary and sufficient to recruit TFE3 onto the lysosomal surface, allowing subsequent phosphorylation of TFE3 by mTORC1. Active mutants of RagC, but not of RagA, rescue both phosphorylation and lysosomal recruitment of TFE3 in the absence of FLCN. These data thus advance the paradigm that mTORC1 substrate specificity is in part conferred by direct recruitment of substrates to the subcellular compartments where mTORC1 resides and identify potential targets for specific modulation of specific branches of the mTOR pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

