Solid Tumor Opioid Receptor Expression and Oncologic Outcomes: Analysis of the Cancer Genome Atlas and Genotype Tissue Expression Project
- PMID: 35359418
- PMCID: PMC8960174
- DOI: 10.3389/fonc.2022.801411
Solid Tumor Opioid Receptor Expression and Oncologic Outcomes: Analysis of the Cancer Genome Atlas and Genotype Tissue Expression Project
Abstract
Background: Opioid receptors are expressed not only by neural cells in the central nervous system, but also by many solid tumor cancer cells. Whether perioperative opioids given for analgesia after tumor resection surgery might inadvertently activate tumor cells, promoting recurrence or metastasis, remains controversial. We analysed large public gene repositories of solid tumors to investigate differences in opioid receptor expression between normal and tumor tissues and their association with long-term oncologic outcomes.
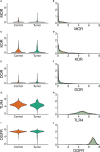
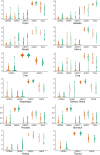
Methods: We investigated the normalized gene expression of µ, κ, δ opioid receptors (MOR, KOR, DOR), Opioid Growth Factor (OGFR), and Toll-Like 4 (TLR4) receptors in normal and tumor samples from twelve solid tumor types. We carried out mixed multivariable logistic and Cox regression analysis on whether there was an association between these receptors' gene expression and the tissue where found, i.e., tumor or normal tissue. We also evaluated the association between tumor opioid receptor gene expression and patient disease-free interval (DFI) and overall survival (OS).
Results: We retrieved 8,780 tissue samples, 5,852 from tumor and 2,928 from normal tissue, of which 2,252 were from the Genotype Tissue Expression Project (GTEx) and 672 from the Cancer Genome Atlas (TCGA) repository. The Odds Ratio (OR) [95%CI] for gene expression of the specific opioid receptors in the examined tumors varied: MOR: 0.74 [0.63-0.87], KOR: 1.27 [1.17-1.37], DOR: 1.66 [1.48-1.87], TLR4: 0.29 [0.26-0.32], OGFR: 2.39 [2.05-2.78]. After controlling all confounding variables, including age and cancer stage, there was no association between tumor opioid receptor expression and long-term oncologic outcomes.
Conclusion: Opioid receptor gene expression varies between different solid tumor types. There was no association between tumor opioid receptor expression and recurrence. Understanding the significance of opioid receptor expression on tumor cells remains elusive.
Keywords: cancer; immunohistochemistry; neoplasm; opioid receptors; perioperative opioid; surgery; tumor.
Copyright © 2022 Belltall, Zúñiga-Trejos, Garrido-Cano, Eroles, Argente-Navarro, Buggy, Díaz-Cambronero and Mazzinari.
Conflict of interest statement
OD-C: Received payment for educational talks and scientific conferences from MSD (Merck Sharp & Dohme, Inc.). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

