A Comprehensive Atlas of Immunological Differences Between Humans, Mice, and Non-Human Primates
- PMID: 35359965
- PMCID: PMC8962947
- DOI: 10.3389/fimmu.2022.867015
A Comprehensive Atlas of Immunological Differences Between Humans, Mice, and Non-Human Primates
Abstract
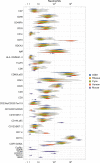
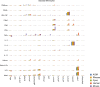
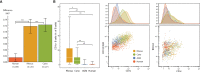
Animal models are an integral part of the drug development and evaluation process. However, they are unsurprisingly imperfect reflections of humans, and the extent and nature of many immunological differences are unknown. With the rise of targeted and biological therapeutics, it is increasingly important that we understand the molecular differences in the immunological behavior of humans and model organisms. However, very few antibodies are raised against non-human primate antigens, and databases of cross-reactivity between species are incomplete. Thus, we screened 332 antibodies in five immune cell populations in blood from humans and four non-human primate species generating a comprehensive cross-reactivity catalog that includes cell type-specificity. We used this catalog to create large mass cytometry universal cross-species phenotyping and signaling panels for humans, along with three of the model organisms most similar to humans: rhesus and cynomolgus macaques and African green monkeys; and one of the mammalian models most widely used in drug development: C57BL/6 mice. As a proof-of-principle, we measured immune cell signaling responses across all five species to an array of 15 stimuli using mass cytometry. We found numerous instances of different cellular phenotypes and immune signaling events occurring within and between species, and detailed three examples (double-positive T cell frequency and signaling; granulocyte response to Bacillus anthracis antigen; and B cell subsets). We also explore the correlation of herpes simian B virus serostatus on the immune profile. Antibody panels and the full dataset generated are available online as a resource to enable future studies comparing immune responses across species during the evaluation of therapeutics.
Keywords: African green monkey (AGM) (Chlorocebus aethiops); CyTOF mass cytometry; cynomolgus monkey (Macaca fascicularis); mouse, immune cell signaling; rhesus macaque (Macaca mulatta).
Copyright © 2022 Bjornson-Hooper, Fragiadakis, Spitzer, Chen, Madhireddy, Hu, Lundsten, McIlwain and Nolan.
Conflict of interest statement
Author KL was employed by BioLegend Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- FDA . US Food and Drug Administration: Animal Rule Information. Available at: https://www.fda.gov/emergency-preparedness-and-response/mcm-regulatory-s... (Accessed December 15, 2021).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

