27-Hydroxycholesterol, The Estrogen Receptor Modulator, Alters DNA Methylation in Breast Cancer
- PMID: 35360070
- PMCID: PMC8961300
- DOI: 10.3389/fendo.2022.783823
27-Hydroxycholesterol, The Estrogen Receptor Modulator, Alters DNA Methylation in Breast Cancer
Abstract
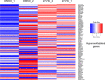
27-hydroxycholesterol (27-HC) is the first known endogenous selective estrogen receptor modulator (SERM), and its elevation from normal levels is closely associated with breast cancer. A plethora of evidence suggests that aberrant epigenetic signatures in breast cancer cells can result in differential responses to various chemotherapeutics and often leads to the development of resistant cancer cells. Such aberrant epigenetic changes are mostly dictated by the microenvironment. The local concentration of oxygen and metabolites in the microenvironment of breast cancer are known to influence the development of breast cancer. Hence, we hypothesized that 27-HC, an oxysterol, which has been shown to induce breast cancer progression via estrogen receptor alpha (ERα) and liver X receptor (LXR) and by modulating immune cells, may also induce epigenetic changes. For deciphering the same, we treated the estrogen receptor-positive cells with 27-HC and identified DNA hypermethylation on a subset of genes by performing DNA bisulfite sequencing. The genes that showed significant DNA hypermethylation were phosphatidylserine synthase 2 (PTDSS2), MIR613, indoleamine 2,3-dioxygenase 1 (IDO1), thyroid hormone receptor alpha (THRA), dystrotelin (DTYN), and mesoderm induction early response 1, family member 3 (MIER). Furthermore, we found that 27-HC weakens the DNMT3B association with the ERα in MCF-7 cells. This study reports that 27-HC induces aberrant DNA methylation changes on the promoters of a subset of genes through modulation of ERα and DNMT3B complexes to induce the local DNA methylation changes, which may dictate drug responses and breast cancer development.
Keywords: 27-hydroxycholesterol; DNA methylation; breast cancer; epigenetics; estrogen receptor alpha; liver X receptor (LXR).
Copyright © 2022 Vini, Rajavelu and Sreeharshan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

