A risk model of gene signatures for predicting platinum response and survival in ovarian cancer
- PMID: 35361267
- PMCID: PMC8973612
- DOI: 10.1186/s13048-022-00969-3
A risk model of gene signatures for predicting platinum response and survival in ovarian cancer
Abstract
Background: Ovarian cancer (OC) is the deadliest tumor in the female reproductive tract. And increased resistance to platinum-based chemotherapy represents the major obstacle in the treatment of OC currently. Robust and accurate gene expression models are crucial tools in distinguishing platinum therapy response and evaluating the prognosis of OC patients.
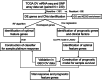
Methods: In this study, 230 samples from The Cancer Genome Atlas (TCGA) OV dataset were subjected to mRNA expression profiling, single nucleotide polymorphism (SNP), and copy number variation (CNV) analysis comprehensively to screen out the differentially expressed genes (DEGs). An SVM classifier and a prognostic model were constructed using the Random Forest algorithm and LASSO Cox regression model respectively via R. The Gene Expression Omnibus (GEO) database was applied as the validation set.
Results: Forty-eight differentially expressed genes (DEGs) were figured out through integrated analysis of gene expression, single nucleotide polymorphism (SNP), and copy number variation (CNV) data. A 10-gene classifier was constructed which could discriminate platinum-sensitive samples precisely with an AUC of 0.971 in the training set and of 0.926 in the GEO dataset (GSE638855). In addition, 8 optimal genes were further selected to construct the prognostic risk model whose predictions were consistent with the actual survival outcomes in the training cohort (p = 9.613e-05) and validated in GSE638855 (p = 0.04862). PNLDC1, SLC5A1, and SYNM were then identified as hub genes that were associated with both platinum response status and prognosis, which was further validated by the Fudan University Shanghai cancer center (FUSCC) cohort.
Conclusion: These findings reveal a specific risk model that could serve as effective biomarkers to identify patients' platinum response status and predict survival outcomes for OC patients. PNLDC1, SLC5A1, and SYNM are the hub genes that may serve as potential biomarkers in OC treatment.
Keywords: Biomarkers; Ovarian cancer; Platinum response; Prognostic model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures






References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Pignata S, Cannella L, Leopardo D, Pisano C, Bruni GS, Facchini G. Chemotherapy in epithelial ovarian cancer. Cancer Lett. 2011;303:73–83. - PubMed
-
- Davis A, Tinker AV, Friedlander M. "platinum resistant" ovarian cancer: what is it, who to treat and how to measure benefit? Gynecol Oncol. 2014;133:624–631. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

