USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy
- PMID: 35361799
- PMCID: PMC8971425
- DOI: 10.1038/s41467-022-29401-6
USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy
Abstract
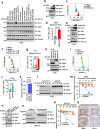
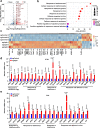
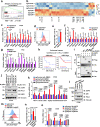
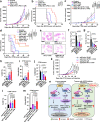
Anti-PD-1/PD-L1 immunotherapy has achieved impressive therapeutic outcomes in patients with multiple cancer types. However, the underlined molecular mechanism(s) for moderate response rate (15-25%) or resistance to PD-1/PD-L1 blockade remains not completely understood. Here, we report that inhibiting the deubiquitinase, USP8, significantly enhances the efficacy of anti-PD-1/PD-L1 immunotherapy through reshaping an inflamed tumor microenvironment (TME). Mechanistically, USP8 inhibition increases PD-L1 protein abundance through elevating the TRAF6-mediated K63-linked ubiquitination of PD-L1 to antagonize K48-linked ubiquitination and degradation of PD-L1. In addition, USP8 inhibition also triggers innate immune response and MHC-I expression largely through activating the NF-κB signaling. Based on these mechanisms, USP8 inhibitor combination with PD-1/PD-L1 blockade significantly activates the infiltrated CD8+ T cells to suppress tumor growth and improves the survival benefit in several murine tumor models. Thus, our study reveals a potential combined therapeutic strategy to utilize a USP8 inhibitor and PD-1/PD-L1 blockade for enhancing anti-tumor efficacy.
© 2022. The Author(s).
Conflict of interest statement
G.J.F. has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, and Novartis. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, and Jubilant. G.J.F. has equity in Nextpoint, Triursus, Xios, and IgM. W.W. is a co-founder and consultant for the ReKindle Therapeutics. Other authors declare no competing interests.
Figures


References
-
- Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

