Macrophages use apoptotic cell-derived methionine and DNMT3A during efferocytosis to promote tissue resolution
- PMID: 35361955
- PMCID: PMC9050866
- DOI: 10.1038/s42255-022-00551-7
Macrophages use apoptotic cell-derived methionine and DNMT3A during efferocytosis to promote tissue resolution
Abstract
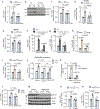
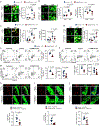
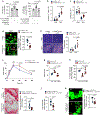
Efferocytosis, the clearance of apoptotic cells (ACs) by macrophages, is critical for tissue resolution, with defects driving many diseases. Mechanisms of efferocytosis-mediated resolution are incompletely understood. Here, we show that AC-derived methionine regulates resolution through epigenetic repression of the extracellular signal-regulated kinase 1/2 (ERK1/2) phosphatase Dusp4. We focus on two key efferocytosis-induced pro-resolving mediators, prostaglandin E2 (PGE2) and transforming growth factor beta 1 (TGF-β1), and show that efferocytosis induces prostaglandin-endoperoxide synthase 2/cyclooxygenase 2 (Ptgs2/COX2), leading to PGE2 synthesis and PGE2-mediated induction of TGF-β1. ERK1/2 phosphorylation/activation by AC-activated CD36 is necessary for Ptgs2 induction, but this is insufficient owing to an ERK-DUSP4 negative feedback pathway that lowers phospho-ERK. However, subsequent AC engulfment and phagolysosomal degradation lead to Dusp4 repression, enabling enhanced p-ERK and induction of the Ptgs2-PGE2-TGF-β1 pathway. Mechanistically, AC-derived methionine is converted to S-adenosylmethionine, which is used by DNA methyltransferase-3A (DNMT3A) to methylate Dusp4. Bone-marrow DNMT3A deletion in mice blocks COX2/PGE2, TGF-β1, and resolution in sterile peritonitis, apoptosis-induced thymus injury and atherosclerosis. Knowledge of how macrophages use AC-cargo and epigenetics to induce resolution provides mechanistic insight and therapeutic options for diseases driven by impaired resolution.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures








Comment in
-
Dying cell-derived SAM switches off inflammation.Nat Metab. 2022 Apr;4(4):414-415. doi: 10.1038/s42255-022-00556-2. Nat Metab. 2022. PMID: 35361956 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM129350/GM/NIGMS NIH HHS/United States
- S10 RR027050/RR/NCRR NIH HHS/United States
- R35 HL145228/HL/NHLBI NIH HHS/United States
- R00 DK115778/DK/NIDDK NIH HHS/United States
- R00 HL145131/HL/NHLBI NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R01 HL127464/HL/NHLBI NIH HHS/United States
- S10 OD030286/OD/NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- T32 HL007343/HL/NHLBI NIH HHS/United States
- R01 HL155431/HL/NHLBI NIH HHS/United States
- P01 HL087123/HL/NHLBI NIH HHS/United States
- K99 HL145131/HL/NHLBI NIH HHS/United States
- S10 OD020056/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

