Updated restraint dictionaries for carbohydrates in the pyranose form
- PMID: 35362468
- PMCID: PMC8972801
- DOI: 10.1107/S2059798322001103
Updated restraint dictionaries for carbohydrates in the pyranose form
Abstract
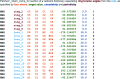
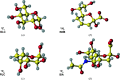
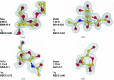
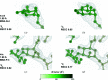
Restraint dictionaries are used during macromolecular structure refinement to encapsulate intramolecular connectivity and geometric information. These dictionaries allow previously determined `ideal' values of features such as bond lengths, angles and torsions to be used as restraint targets. During refinement, restraints influence the model to adopt a conformation that agrees with prior observation. This is especially important when refining crystal structures of glycosylated proteins, as their resolutions tend to be worse than those of nonglycosylated proteins. Pyranosides, the overwhelming majority component in all forms of protein glycosylation, often display conformational errors in crystal structures. Whilst many of these flaws usually relate to model building, refinement issues may also have their root in suboptimal restraint dictionaries. In order to avoid subsequent misinterpretation and to improve the quality of all pyranose monosaccharide entries in the CCP4 Monomer Library, new dictionaries with improved ring torsion restraints, coordinates reflecting the lowest-energy ring pucker and updated geometry have been produced and evaluated. These new dictionaries are now part of the CCP4 Monomer Library and will be released with CCP4 version 8.0.
Keywords: carbohydrates; dictionaries; pyranose; restraints; ring conformation.
open access.
Figures



References
-
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nat. Chem. Biol. 11, 303. - PubMed
-
- Agirre, J., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2017). Curr. Opin. Struct. Biol. 44, 39–47. - PubMed
-
- Agirre, J., Iglesias-Fernández, J., Rovira, C., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2015). Nat. Struct. Mol. Biol. 22, 833–834. - PubMed
-
- Atanasova, M., Bagdonas, H. & Agirre, J. (2020). Curr. Opin. Struct. Biol. 62, 70–78. - PubMed

