μMap-Red: Proximity Labeling by Red Light Photocatalysis
- PMID: 35363468
- PMCID: PMC9843638
- DOI: 10.1021/jacs.2c01384
μMap-Red: Proximity Labeling by Red Light Photocatalysis
Abstract
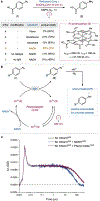
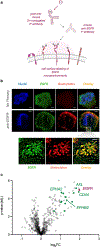
Modern proximity labeling techniques have enabled significant advances in understanding biomolecular interactions. However, current tools primarily utilize activation modes that are incompatible with complex biological environments, limiting our ability to interrogate cell- and tissue-level microenvironments in animal models. Here, we report μMap-Red, a proximity labeling platform that uses a red-light-excited SnIV chlorin e6 catalyst to activate a phenyl azide biotin probe. We validate μMap-Red by demonstrating photonically controlled protein labeling in vitro through several layers of tissue, and we then apply our platform in cellulo to label EGFR microenvironments and validate performance with STED microscopy and quantitative proteomics. Finally, to demonstrate labeling in a complex biological sample, we deploy μMap-Red in whole mouse blood to profile erythrocyte cell-surface proteins. This work represents a significant methodological advance toward light-based proximity labeling in complex tissue environments and animal models.
Figures



References
-
- Keskin O; Tuncbag N; Gursoy A, Predicting protein–protein interactions from the molecular to the proteome level. Chem. Rev, 2016, 116 (8), 4884–4909. - PubMed
-
- Hentze MW; Castello A; Schwarzl T; Preiss T, A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell. Biol, 2018, 19 (5), 327–341. - PubMed
-
- Seath CP; Trowbridge AD; Muir TW; MacMillan DW, Reactive intermediates for interactome mapping. Chem. Soc. Rev, 2021, 50,2911–2926. - PubMed
-
- Rees JS; Li XW; Perrett S; Lilley KS; Jackson AP, Selective proteomic proximity labeling assay using tyramide (SPPLAT): a quantitative method for the proteomic analysis of localized membrane‐bound protein clusters. Curr. Protoc. Protein Sci, 2017, 88 (1), 19.27. 1–19.27. 18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

