Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle
- PMID: 35363556
- PMCID: PMC9169775
- DOI: 10.1073/pnas.2119467119
Structural conservation among variants of the SARS-CoV-2 spike postfusion bundle
Abstract
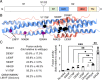
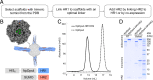
Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) challenge currently available COVID-19 vaccines and monoclonal antibody therapies due to structural and dynamic changes of the viral spike glycoprotein (S). The heptad repeat 1 (HR1) and heptad repeat 2 (HR2) domains of S drive virus–host membrane fusion by assembly into a six-helix bundle, resulting in delivery of viral RNA into the host cell. We surveyed mutations of currently reported SARS-CoV-2 variants and selected eight mutations, including Q954H, N969K, and L981F from the Omicron variant, in the postfusion HR1HR2 bundle for functional and structural studies. We designed a molecular scaffold to determine cryogenic electron microscopy (cryo-EM) structures of HR1HR2 at 2.2–3.8 Å resolution by linking the trimeric N termini of four HR1 fragments to four trimeric C termini of the Dps4 dodecamer from Nostoc punctiforme. This molecular scaffold enables efficient sample preparation and structure determination of the HR1HR2 bundle and its mutants by single-particle cryo-EM. Our structure of the wild-type HR1HR2 bundle resolves uncertainties in previously determined structures. The mutant structures reveal side-chain positions of the mutations and their primarily local effects on the interactions between HR1 and HR2. These mutations do not alter the global architecture of the postfusion HR1HR2 bundle, suggesting that the interfaces between HR1 and HR2 are good targets for developing antiviral inhibitors that should be efficacious against all known variants of SARS-CoV-2 to date. We also note that this work paves the way for similar studies in more distantly related viruses.
Keywords: COVID-19; HR1HR2; SARS-CoV-2; cryogenic electron microscopy; membrane fusion.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Manzanares-Meza L. D., Medina-Contreras O., SARS-CoV-2 and influenza: A comparative overview and treatment implications. Bol. Méd. Hosp. Infant. México 77, 262–273 (2020). - PubMed
-
- Wang P., et al. , Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). - PubMed
-
- Wibmer C. K., et al. , SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 27, 622–625 (2021). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

