Matrix Inversion and Subset Selection (MISS): A pipeline for mapping of diverse cell types across the murine brain
- PMID: 35363567
- PMCID: PMC9168512
- DOI: 10.1073/pnas.2111786119
Matrix Inversion and Subset Selection (MISS): A pipeline for mapping of diverse cell types across the murine brain
Abstract
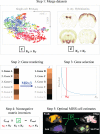
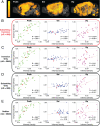
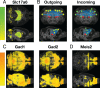
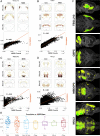
The advent of increasingly sophisticated imaging platforms has allowed for the visualization of the murine nervous system at single-cell resolution. However, current experimental approaches have not yet produced whole-brain maps of a comprehensive set of neuronal and nonneuronal types that approaches the cellular diversity of the mammalian cortex. Here, we aim to fill in this gap in knowledge with an open-source computational pipeline, Matrix Inversion and Subset Selection (MISS), that can infer quantitatively validated distributions of diverse collections of neural cell types at 200-μm resolution using a combination of single-cell RNA sequencing (RNAseq) and in situ hybridization datasets. We rigorously demonstrate the accuracy of MISS against literature expectations. Importantly, we show that gene subset selection, a procedure by which we filter out low-information genes prior to performing deconvolution, is a critical preprocessing step that distinguishes MISS from its predecessors and facilitates the production of cell-type maps with significantly higher accuracy. We also show that MISS is generalizable by generating high-quality cell-type maps from a second independently curated single-cell RNAseq dataset. Together, our results illustrate the viability of computational approaches for determining the spatial distributions of a wide variety of cell types from genetic data alone.
Keywords: cell-type maps; deconvolution; neuroanatomy; transcriptomics.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

