Cross-population selection signatures in Canchim composite beef cattle
- PMID: 35363779
- PMCID: PMC8975110
- DOI: 10.1371/journal.pone.0264279
Cross-population selection signatures in Canchim composite beef cattle
Abstract
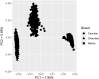
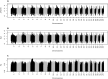
Analyses of livestock genomes have been used to detect selection signatures, which are genomic regions associated with traits under selection leading to a change in allele frequency. The objective of the present study was to characterize selection signatures in Canchim composite beef cattle using cross-population analyses with the founder Nelore and Charolais breeds. High-density single nucleotide polymorphism genotypes were available on 395 Canchim representing the target population, along with genotypes from 809 Nelore and 897 Charolais animals representing the reference populations. Most of the selection signatures were co-located with genes whose functions agree with the expectations of the breeding programs; these genes have previously been reported to associate with meat quality, as well as reproductive traits. Identified genes were related to immunity, adaptation, morphology, as well as behavior, could give new perspectives for understanding the genetic architecture of Canchim. Some selection signatures identified genes that were recently introduced in Canchim, such as the loci related to the polled trait.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Alencar MM. Bovino—Raça Canchim: Origem e Desenvolvimento. Brasília: Embrapa-DMU; 1988.
-
- Wright S. The Genetical Structure of Populations. Annals of Eugenics. 1949;15: 323–354. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

