Chromosome-Level Genome Assembly Reveals Dynamic Sex Chromosomes in Neotropical Leaf-Litter Geckos (Sphaerodactylidae: Sphaerodactylus)
- PMID: 35363859
- PMCID: PMC9270867
- DOI: 10.1093/jhered/esac016
Chromosome-Level Genome Assembly Reveals Dynamic Sex Chromosomes in Neotropical Leaf-Litter Geckos (Sphaerodactylidae: Sphaerodactylus)
Abstract
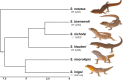
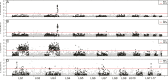
Sex determination is a critical element of successful vertebrate development, suggesting that sex chromosome systems might be evolutionarily stable across lineages. For example, mammals and birds have maintained conserved sex chromosome systems over long evolutionary time periods. Other vertebrates, in contrast, have undergone frequent sex chromosome transitions, which is even more amazing considering we still know comparatively little across large swaths of their respective phylogenies. One reptile group in particular, the gecko lizards (infraorder Gekkota), shows an exceptional lability with regard to sex chromosome transitions and may possess the majority of transitions within squamates (lizards and snakes). However, detailed genomic and cytogenetic information about sex chromosomes is lacking for most gecko species, leaving large gaps in our understanding of the evolutionary processes at play. To address this, we assembled a chromosome-level genome for a gecko (Sphaerodactylidae: Sphaerodactylus) and used this assembly to search for sex chromosomes among six closely related species using a variety of genomic data, including whole-genome re-sequencing, RADseq, and RNAseq. Previous work has identified XY systems in two species of Sphaerodactylus geckos. We expand upon that work to identify between two and four sex chromosome cis-transitions (XY to a new XY) within the genus. Interestingly, we confirmed two different linkage groups as XY sex chromosome systems that were previously unknown to act as sex chromosomes in tetrapods (syntenic with Gallus chromosome 3 and Gallus chromosomes 18/30/33), further highlighting a unique and fascinating trend that most linkage groups have the potential to act as sex chromosomes in squamates.
Keywords: genome evolution; genomics; herpetology; sex chromosomes; sex determination.
© The Author(s) 2022. Published by Oxford University Press on behalf of The American Genetic Association. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




References
-
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

