Inhibition of SARS-CoV-2 replication in the lung with siRNA/VIPER polyplexes
- PMID: 35364120
- PMCID: PMC8963978
- DOI: 10.1016/j.jconrel.2022.03.051
Inhibition of SARS-CoV-2 replication in the lung with siRNA/VIPER polyplexes
Abstract
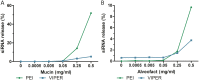
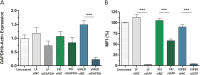
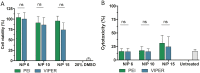
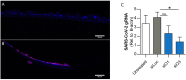
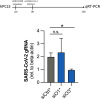
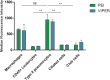
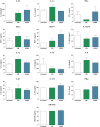
SARS-CoV-2 has been the cause of a global pandemic since 2019 and remains a medical urgency. siRNA-based therapies are a promising strategy to fight viral infections. By targeting a specific region of the viral genome, siRNAs can efficiently downregulate viral replication and suppress viral infection. However, to achieve the desired therapeutic activity, siRNA requires a suitable delivery system. The VIPER (virus-inspired polymer for endosomal release) block copolymer has been reported as promising delivery system for both plasmid DNA and siRNA in the past years. It is composed of a hydrophilic block for condensation of nucleic acids as well as a hydrophobic, pH-sensitive block that, at acidic pH, exposes the membrane lytic peptide melittin, which enhances endosomal escape. In this study, we aimed at developing a formulation for pulmonary administration of siRNA to suppress SARS-CoV-2 replication in lung epithelial cells. After characterizing siRNA/VIPER polyplexes, the activity and safety profile were confirmed in a lung epithelial cell line. To further investigate the activity of the polyplexes in a more sophisticated cell culture system, an air-liquid interface (ALI) culture was established. siRNA/VIPER polyplexes reached the cell monolayer and penetrated through the mucus layer secreted by the cells. Additionally, the activity against wild-type SARS-CoV-2 in the ALI model was confirmed by qRT-PCR. To investigate translatability of our findings, the activity against SARS-CoV-2 was tested ex vivo in human lung explants. Here, siRNA/VIPER polyplexes efficiently inhibited SARS-CoV-2 replication. Finally, we verified the delivery of siRNA/VIPER polyplexes to lung epithelial cells in vivo, which represent the main cellular target of viral infection in the lung. In conclusion, siRNA/VIPER polyplexes efficiently delivered siRNA to lung epithelial cells and mediated robust downregulation of viral replication both in vitro and ex vivo without toxic or immunogenic side effects in vivo, demonstrating the potential of local siRNA delivery as a promising antiviral therapy in the lung.
Keywords: Human precision-cut lung slices; Pulmonary delivery; RNA therapeutics; SARS-CoV-2; siRNA delivery.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
T.M. is an
Figures

References
-
- World Health Organization 2021. https://covid19.who.int/https://covid19.who.int/
-
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/nejmoa2002032. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

