The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases
- PMID: 35365203
- PMCID: PMC8973751
- DOI: 10.1186/s12920-022-01216-w
The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases
Abstract
Background: The remarkable growth of genome-wide association studies (GWAS) has created a critical need to experimentally validate the disease-associated variants, 90% of which involve non-coding variants.
Methods: To determine how the field is addressing this urgent need, we performed a comprehensive literature review identifying 36,676 articles. These were reduced to 1454 articles through a set of filters using natural language processing and ontology-based text-mining. This was followed by manual curation and cross-referencing against the GWAS catalog, yielding a final set of 286 articles.
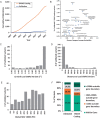
Results: We identified 309 experimentally validated non-coding GWAS variants, regulating 252 genes across 130 human disease traits. These variants covered a variety of regulatory mechanisms. Interestingly, 70% (215/309) acted through cis-regulatory elements, with the remaining through promoters (22%, 70/309) or non-coding RNAs (8%, 24/309). Several validation approaches were utilized in these studies, including gene expression (n = 272), transcription factor binding (n = 175), reporter assays (n = 171), in vivo models (n = 104), genome editing (n = 96) and chromatin interaction (n = 33).
Conclusions: This review of the literature is the first to systematically evaluate the status and the landscape of experimentation being used to validate non-coding GWAS-identified variants. Our results clearly underscore the multifaceted approach needed for experimental validation, have practical implications on variant prioritization and considerations of target gene nomination. While the field has a long way to go to validate the thousands of GWAS associations, we show that progress is being made and provide exemplars of validation studies covering a wide variety of mechanisms, target genes, and disease areas.
Keywords: Experimental validation; Functional variant; GWAS; Non-coding; Systematic review.
© 2022. The Author(s).
Conflict of interest statement
AA, SW, EK, JR, SG, ST and HJ are employees of AbbVie. LS, JWD and JL were employees of AbbVie at the time of the study.
Figures



References
-
- Collins FS, Doudna JA, Lander ES, Rotimi CN. Human molecular genetics and genomics—Important advances and exciting possibilities. N Engl J Med. 2021;384:1–4. - PubMed
-
- OMIM - Online Mendelian Inheritance in Man. https://www.omim.org/. 2021 [cited 2021 Apr 11]; Available from: https://www.omim.org/
-
- Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

