Surveillance of Enterobacter cloacae complex colonization and comparative analysis of different typing methods on a neonatal intensive care unit in Germany
- PMID: 35365217
- PMCID: PMC8973561
- DOI: 10.1186/s13756-022-01094-y
Surveillance of Enterobacter cloacae complex colonization and comparative analysis of different typing methods on a neonatal intensive care unit in Germany
Abstract
Background: Enterobacter cloacae complex is a group of common opportunistic pathogens on neonatal intensive care units. Active microbiological screening to guide empirical antimicrobial treatment or to detect transmission events is recommended in high-risk preterm neonates. A rise in colonization with E. cloacae complex was observed in a German perinatal centre. The aim of this study was to evaluate the performance of different typing techniques using whole genome sequencing (WGS) as a reference.
Methods: Enterobacter cloacae complex isolates from clinical and screening specimens with an epidemiological link to the neonatal intensive care units were further assessed. Identification and antibiotic susceptibility testing was performed by a combination of VITEK2 (bioMérieux) and MALDI-TOF (Bruker Daltonics), followed by RAPD/rep-PCR and PFGE (XbaI). Retrospectively, all isolates were analyzed by Fourier-transform infrared (FTIR) spectroscopy (IR Biotyper, Bruker Daltonics). Whole genome sequencing with SNP-based clustering was used as the reference method. Furthermore, resistome analysis, sequence type and species identification were derived from the WGS data. Transmission analysis was based on epidemiological and typing data.
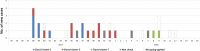
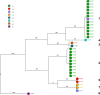
Results: Between September 2017 and March 2018 32 mostly preterm neonates were found to be colonized with E. cloacae complex and 32 isolates from 24 patients were available for further typing. RAPD/rep-PCR and PFGE showed good concordance with WGS whereas FTIR displayed mediocre results [adjusted rand index (ARI) = 0.436]. A polyclonal increase and two dominant and overlapping clonal clusters of two different E. hormaechei subspecies were detected. Overall, four different species were identified. Genotyping confirmed third-generation cephalosporin resistance development in isolates of the same patient. During the six-month period several infection prevention interventions were performed and no E. cloacae complex isolates were observed during the following months.
Conclusions: Interpretation of the microbiological results alone to detect transmission events is often challenging and bacterial typing is of utmost importance to implement targeted infection control measures in an epidemic occurrence of E. cloacae complex. WGS is the most discriminatory method. However, traditional methods such as PFGE or RAPD/rep-PCR can provide reliable and quicker results in many settings. Furthermore, research is needed to quickly identify E. cloacae complex to the species level in the microbiological laboratory.
Keywords: Bacterial typing; E. cloacae complex; Neonatal colonization screening; Neonatal intensive care unit.
© 2022. The Author(s).
Conflict of interest statement
All authors have no conflicts of interest.
Figures
References
-
- Kremer A, Hoffmann H. Prevalences of the Enterobacter cloacae complex and its phylogenetic derivatives in the nosocomial environment. Eur J Clin Microbiol Infect Dis. 2012;31:2951–2955. - PubMed
-
- Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

