Clinical and translational values of spatial transcriptomics
- PMID: 35365599
- PMCID: PMC8972902
- DOI: 10.1038/s41392-022-00960-w
Clinical and translational values of spatial transcriptomics
Abstract
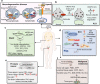
The combination of spatial transcriptomics (ST) and single cell RNA sequencing (scRNA-seq) acts as a pivotal component to bridge the pathological phenomes of human tissues with molecular alterations, defining in situ intercellular molecular communications and knowledge on spatiotemporal molecular medicine. The present article overviews the development of ST and aims to evaluate clinical and translational values for understanding molecular pathogenesis and uncovering disease-specific biomarkers. We compare the advantages and disadvantages of sequencing- and imaging-based technologies and highlight opportunities and challenges of ST. We also describe the bioinformatics tools necessary on dissecting spatial patterns of gene expression and cellular interactions and the potential applications of ST in human diseases for clinical practice as one of important issues in clinical and translational medicine, including neurology, embryo development, oncology, and inflammation. Thus, clear clinical objectives, designs, optimizations of sampling procedure and protocol, repeatability of ST, as well as simplifications of analysis and interpretation are the key to translate ST from bench to clinic.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Wang X. Clinical trans-omics: an integration of clinical phenomes with molecular multiomics. Cell Biol. Toxicol. 2018;34:163–166. - PubMed
-
- Liao J, Lu X, Shao X, Zhu L, Fan X. Uncovering an organ’s molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2021;39:43–58. - PubMed
-
- Stahl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

