Imaging vesicle formation dynamics supports the flexible model of clathrin-mediated endocytosis
- PMID: 35365614
- PMCID: PMC8976038
- DOI: 10.1038/s41467-022-29317-1
Imaging vesicle formation dynamics supports the flexible model of clathrin-mediated endocytosis
Abstract
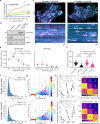
Clathrin polymerization and changes in plasma membrane architecture are necessary steps in forming vesicles to internalize cargo during clathrin-mediated endocytosis (CME). Simultaneous analysis of clathrin dynamics and membrane structure is challenging due to the limited axial resolution of fluorescence microscopes and the heterogeneity of CME. This has fueled conflicting models of vesicle assembly and obscured the roles of flat clathrin assemblies. Here, using Simultaneous Two-wavelength Axial Ratiometry (STAR) microscopy, we bridge this critical knowledge gap by quantifying the nanoscale dynamics of clathrin-coat shape change during vesicle assembly. We find that de novo clathrin accumulations generate both flat and curved structures. High-throughput analysis reveals that the initiation of vesicle curvature does not directly correlate with clathrin accumulation. We show clathrin accumulation is preferentially simultaneous with curvature formation at shorter-lived clathrin-coated vesicles (CCVs), but favors a flat-to-curved transition at longer-lived CCVs. The broad spectrum of curvature initiation dynamics revealed by STAR microscopy supports multiple productive mechanisms of vesicle formation and advocates for the flexible model of CME.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

