Long-chain ceramides are cell non-autonomous signals linking lipotoxicity to endoplasmic reticulum stress in skeletal muscle
- PMID: 35365625
- PMCID: PMC8975934
- DOI: 10.1038/s41467-022-29363-9
Long-chain ceramides are cell non-autonomous signals linking lipotoxicity to endoplasmic reticulum stress in skeletal muscle
Abstract
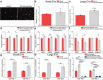
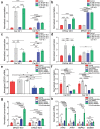
The endoplasmic reticulum (ER) regulates cellular protein and lipid biosynthesis. ER dysfunction leads to protein misfolding and the unfolded protein response (UPR), which limits protein synthesis to prevent cytotoxicity. Chronic ER stress in skeletal muscle is a unifying mechanism linking lipotoxicity to metabolic disease. Unidentified signals from cells undergoing ER stress propagate paracrine and systemic UPR activation. Here, we induce ER stress and lipotoxicity in myotubes. We observe ER stress-inducing lipid cell non-autonomous signal(s). Lipidomics identifies that palmitate-induced cell stress induces long-chain ceramide 40:1 and 42:1 secretion. Ceramide synthesis through the ceramide synthase 2 de novo pathway is regulated by UPR kinase Perk. Inactivation of CerS2 in mice reduces systemic and muscle ceramide signals and muscle UPR activation. The ceramides are packaged into extracellular vesicles, secreted and induce UPR activation in naïve myotubes through dihydroceramide accumulation. This study furthers our understanding of ER stress by identifying UPR-inducing cell non-autonomous signals.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

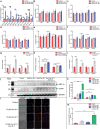
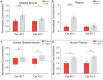
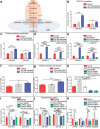

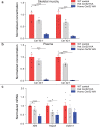
References
-
- Palomer X, Pizarro-Delgado J, Barroso E, Vazquez-Carrera M. Palmitic and Oleic Acid: The Yin and Yang of Fatty Acids in Type 2 Diabetes Mellitus. Trends Endocrinol. Metab. 2018;29:178–190. - PubMed
-
- Unger RH, Clark GO, Scherer PE, Orci L. Lipid homeostasis, lipotoxicity and the metabolic syndrome. Biochim. Biophys. Acta. 2010;1801:209–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/R013500/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 16/0005382/DUK_/Diabetes UK/United Kingdom
- BB/T004231/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00014/5/MRC_/Medical Research Council/United Kingdom
- MR/R014086/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

