A functional map of HIV-host interactions in primary human T cells
- PMID: 35365639
- PMCID: PMC8976027
- DOI: 10.1038/s41467-022-29346-w
A functional map of HIV-host interactions in primary human T cells
Abstract
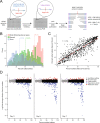
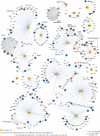
Human Immunodeficiency Virus (HIV) relies on host molecular machinery for replication. Systematic attempts to genetically or biochemically define these host factors have yielded hundreds of candidates, but few have been functionally validated in primary cells. Here, we target 426 genes previously implicated in the HIV lifecycle through protein interaction studies for CRISPR-Cas9-mediated knock-out in primary human CD4+ T cells in order to systematically assess their functional roles in HIV replication. We achieve efficient knockout (>50% of alleles) in 364 of the targeted genes and identify 86 candidate host factors that alter HIV infection. 47 of these factors validate by multiplex gene editing in independent donors, including 23 factors with restrictive activity. Both gene editing efficiencies and HIV-1 phenotypes are highly concordant among independent donors. Importantly, over half of these factors have not been previously described to play a functional role in HIV replication, providing numerous novel avenues for understanding HIV biology. These data further suggest that host-pathogen protein-protein interaction datasets offer an enriched source of candidates for functional host factor discovery and provide an improved understanding of the mechanics of HIV replication in primary T cells.
© 2022. The Author(s).
Conflict of interest statement
A.M. is a compensated cofounder, member of the boards of directors, and a member of the scientific advisory boards of Spotlight Therapeutics and Arsenal Biosciences. A.M. is a cofounder, member of the boards of directors, and a member of the scientific advisory board of Survey Genomics. A.M. is a compensated member of the scientific advisory board of NewLimit. A.M. owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, PACT Pharma, and Merck. AM has received fees from 23andMe, PACT Pharma, Juno Therapeutics, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein, and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. The Marson lab has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead, and Anthem. N.J.K. has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics, and Interline Therapeutics. He is a shareholder in Tenaya Therapeutics, Maze Therapeutics and Interline Therapeutics, and is financially compensated by GEn1E Lifesciences, Inc. and Twist Bioscience Corp. The Krogan Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche.
Figures




References
-
- Richman DD, et al. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AI165236/AI/NIAID NIH HHS/United States
- R01 AI120694/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K22 AI136691/AI/NIAID NIH HHS/United States
- R01 AI150998/AI/NIAID NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- P50 AI150476/AI/NIAID NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- T32 GM141323/GM/NIGMS NIH HHS/United States
- P30 AI117943/AI/NIAID NIH HHS/United States
- DP2 DA042423/DA/NIDA NIH HHS/United States
- U54 AI170792/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

