A high-quality assembled genome and its comparative analysis decode the adaptive molecular mechanism of the number one Chinese cotton variety CRI-12
- PMID: 35365835
- PMCID: PMC8975723
- DOI: 10.1093/gigascience/giac019
A high-quality assembled genome and its comparative analysis decode the adaptive molecular mechanism of the number one Chinese cotton variety CRI-12
Abstract
Background: Gossypium hirsutum L. is the most widely cultivated cotton species, and a high-quality reference genome would be a huge boost for researching the molecular mechanism of agronomic traits in cotton.
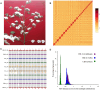
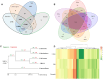
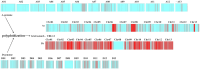
Findings: Here, Pacific Biosciences and Hi-C sequencing technologies were used to assemble a new upland cotton genome of the No. 1 Chinese cotton variety CRI-12. We generated a high-quality assembled CRI-12 genome of 2.31 Gb with a contig N50 of 19.65 Mb, which was superior to previously reported genomes. Comparisons between CRI-12 and other reported genomes revealed 7,966 structural variations and 7,378 presence/absence variations. The distribution of the haplotypes among A-genome (Gossypium arboreum), D-genome (Gossypium raimondii), and AD-genome (G. hirsutum and Gossypium barbadense) suggested that many haplotypes were lost and recombined in the process of polyploidization. More than half of the haplotypes that correlated with different tolerances were located on chromosome D13, suggesting that this chromosome may be important for wide adaptation. Finally, it was demonstrated that DNA methylation may provide advantages in environmental adaptation through whole-genome bisulfite sequencing analysis.
Conclusions: This research provides a new reference genome for molecular biology research on Gossypium hirsutum L. and helps decode the broad environmental adaptation mechanisms in the No. 1 Chinese cotton variety CRI-12.
Keywords: CRI-12; DNA methylation; annotation; genome assembly; haplotypes.
© The Author(s) 2022. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



Similar articles
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Extensive haplotypes are associated with population differentiation and environmental adaptability in Upland cotton (Gossypium hirsutum).Theor Appl Genet. 2020 Dec;133(12):3273-3285. doi: 10.1007/s00122-020-03668-z. Epub 2020 Aug 25. Theor Appl Genet. 2020. PMID: 32844253
-
Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid Upland cotton (Gossypium hirsutum L.).BMC Genomics. 2013 Mar 28;14:208. doi: 10.1186/1471-2164-14-208. BMC Genomics. 2013. PMID: 23537070 Free PMC article.
-
High-quality Gossypium hirsutum and Gossypium barbadense genome assemblies reveal the landscape and evolution of centromeres.Plant Commun. 2024 Feb 12;5(2):100722. doi: 10.1016/j.xplc.2023.100722. Epub 2023 Sep 22. Plant Commun. 2024. PMID: 37742072 Free PMC article.
-
Cotton2035: From genomics research to optimized breeding.Mol Plant. 2025 Feb 3;18(2):298-312. doi: 10.1016/j.molp.2025.01.010. Epub 2025 Jan 21. Mol Plant. 2025. PMID: 39844464 Review.
Cited by
-
A chromosome-level reference genome of a Convolvulaceae species Ipomoea cairica.G3 (Bethesda). 2022 Aug 25;12(9):jkac187. doi: 10.1093/g3journal/jkac187. G3 (Bethesda). 2022. PMID: 35894697 Free PMC article.
-
Cotton pedigree genome reveals restriction of cultivar-driven strategy in cotton breeding.Genome Biol. 2023 Dec 8;24(1):282. doi: 10.1186/s13059-023-03124-3. Genome Biol. 2023. PMID: 38066616 Free PMC article.
-
Impacts of parental genomic divergence in non-syntenic regions on cotton heterosis.J Adv Res. 2025 Jul;73:15-29. doi: 10.1016/j.jare.2024.08.010. Epub 2024 Aug 6. J Adv Res. 2025. PMID: 39111623 Free PMC article.
-
Systematic identification of TPS genes in Gossypium and their characteristics in response to flooding stress.Front Plant Sci. 2023 Feb 8;14:1126884. doi: 10.3389/fpls.2023.1126884. eCollection 2023. Front Plant Sci. 2023. PMID: 36844072 Free PMC article.
-
Pangenome analysis reveals yield- and fiber-related diversity and interspecific gene flow in Gossypium barbadense L.Nat Commun. 2025 May 29;16(1):4995. doi: 10.1038/s41467-025-60254-x. Nat Commun. 2025. PMID: 40442108 Free PMC article.
References
-
- Hu Y, Chen J, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–48. - PubMed
-
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8(2):135–41. - PubMed
-
- Paterson AH, Wendel JF, Gundlach H, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–7. - PubMed
-
- Wang M, Tu L, Lin M, et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet. 2017;49(4):579–87. - PubMed
-
- Zhang T, Hu Y, Jiang W, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

