Mechanisms of action of triptolide against colorectal cancer: insights from proteomic and phosphoproteomic analyses
- PMID: 35366242
- PMCID: PMC9037262
- DOI: 10.18632/aging.203992
Mechanisms of action of triptolide against colorectal cancer: insights from proteomic and phosphoproteomic analyses
Abstract
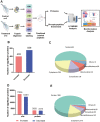
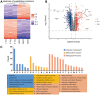
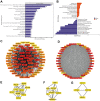
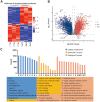
Triptolide is a potent anti-inflammatory agent that also possesses anticancer activity, including against colorectal cancer (CRC), one of the most frequent cancers around the world. In order to clarify how triptolide may be effective against CRC, we analyzed the proteome and phosphoproteome of CRC cell line HCT116 after incubation for 48 h with the drug (40 nM) or vehicle. Tandem mass tagging led to the identification of 403 proteins whose levels increased and 559 whose levels decreased in the presence of triptolide. We also identified 3,110 sites in proteins that were phosphorylated at higher levels and 3,161 sites phosphorylated at lower levels in the presence of the drug. Analysis of these differentially expressed and/or phosphorylated proteins showed that they were enriched in pathways involving ribosome biogenesis, PI3K-Akt signaling, MAPK signaling, nucleic acid binding as well as other pathways. Protein-protein interactions were explored using the STRING database, and we identified nine protein modules and 15 hub proteins. Finally, we identified 57 motifs using motif analysis of phosphosites and found 16 motifs were experimentally verified for known protein kinases, while 41 appear to be novel. These findings may help clarify how triptolide works against CRC and may guide the development of novel treatments.
Keywords: colorectal cancer; molecular docking; phosphoproteomic; proteomic; triptolide.
Conflict of interest statement
Figures


References
-
- Oliveira A, Beyer G, Chugh R, Skube SJ, Majumder K, Banerjee S, Sangwan V, Li L, Dawra R, Subramanian S, Saluja A, Dudeja V. Triptolide abrogates growth of colon cancer and induces cell cycle arrest by inhibiting transcriptional activation of E2F. Lab Invest. 2015; 95:648–59. 10.1038/labinvest.2015.46 - DOI - PMC - PubMed
-
- Acikgoz E, Tatar C, Oktem G. Triptolide inhibits CD133+ /CD44+ colon cancer stem cell growth and migration through triggering apoptosis and represses epithelial-mesenchymal transition via downregulating expressions of snail, slug, and twist. J Cell Biochem. 2020; 121:3313–24. 10.1002/jcb.29602 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

