bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum
- PMID: 35366869
- PMCID: PMC8977021
- DOI: 10.1186/s12934-022-01765-w
bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum
Abstract
Background: Reactive oxygen species (ROS) trigger different morphogenic processes in filamentous fungi and have been shown to play a role in the regulation of the biosynthesis of some secondary metabolites. Some bZIP transcription factors, such as Yap1, AtfA and AtfB, mediate resistance to oxidative stress and have a role in secondary metabolism regulation. In this work we aimed to get insight into the molecular basis of this regulation in the industrially important fungus Penicillium chrysogenum through the characterization of the role played by two effectors that mediate the oxidative stress response in development and secondary metabolism.
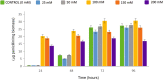
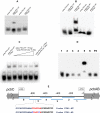
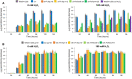
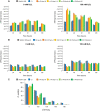
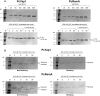
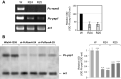
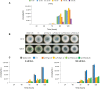
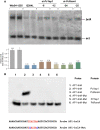
Results: In P. chrysogenum, penicillin biosynthesis and conidiation are stimulated by the addition of H2O2 to the culture medium, and this effect is mediated by the bZIP transcription factors PcYap1 and PcRsmA. Silencing of expression of both proteins by RNAi resulted in similar phenotypes, characterized by increased levels of ROS in the cell, reduced conidiation, higher sensitivity of conidia to H2O2 and a decrease in penicillin production. Both PcYap1 and PcRsmA are able to sense H2O2-generated ROS in vitro and change its conformation in response to this stimulus. PcYap1 and PcRsmA positively regulate the expression of brlA, the first gene of the conidiation central regulatory pathway. PcYap1 binds in vitro to a previously identified regulatory sequence in the promoter of the penicillin gene pcbAB: TTAGTAA, and to a TTACTAA sequence in the promoter of the brlA gene, whereas PcRsmA binds to the sequences TGAGACA and TTACGTAA (CRE motif) in the promoters of the pcbAB and penDE genes, respectively.
Conclusions: bZIP transcription factors PcYap1 and PcRsmA respond to the presence of H2O2-generated ROS and regulate oxidative stress response in the cell. Both proteins mediate ROS regulation of penicillin biosynthesis and conidiation by binding to specific regulatory elements in the promoters of key genes. PcYap1 is identified as the previously proposed transcription factor PTA1 (Penicillin Transcriptional Activator 1), which binds to the regulatory sequence TTAGTAA in the pcbAB gene promoter. This is the first report of a Yap1 protein directly regulating transcription of a secondary metabolism gene. A model describing the regulatory network mediated by PcYap1 and PcRsmA is proposed.
Keywords: DNA-binding proteins; Fungal morphogenesis; Oxidative stress defense; Reactive oxygen species; RsmA; Secondary metabolism; Transcriptional regulation; Yap1; bZIP transcription factor.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

