Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant
- PMID: 35367284
- PMCID: PMC8968180
- DOI: 10.1016/j.virusres.2022.198765
Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant
Abstract
Background: Emergence of new variant of SARS-CoV-2, namely omicron, has posed a global concern because of its high rate of transmissibility and mutations in its genome. Researchers worldwide are trying to understand the evolution and emergence of such variants to understand the mutational cascade events.
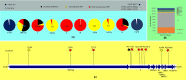
Methods: We have considered all omicron genomes (n = 302 genomes) available till 2nd December 2021 in the public repository of GISAID along with representatives of variants of concern (VOC), i.e., alpha, beta, gamma, delta, and omicron; variant of interest (VOI) mu and lambda; and variant under monitoring (VUM). Whole genome-based phylogeny and mutational analysis were performed to understand the evolution of SARS CoV-2 leading to emergence of omicron variant.
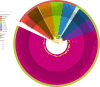
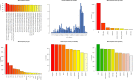
Results: Whole genome-based phylogeny depicted two phylogroups (PG-I and PG-II) forming variant specific clades except for gamma and VUM GH. Mutational analysis detected 18,261 mutations in the omicron variant, majority of which were non-synonymous mutations in spike (A67, T547K, D614G, H655Y, N679K, P681H, D796Y, N856K, Q954H), followed by RNA dependent RNA polymerase (rdrp) (A1892T, I189V, P314L, K38R, T492I, V57V), ORF6 (M19M) and nucleocapsid protein (RG203KR).
Conclusion: Delta and omicron have evolutionary diverged into distinct phylogroups and do not share a common ancestry. While, omicron shares common ancestry with VOI lambda and its evolution is mainly derived by the non-synonymous mutations.
Keywords: NSP, non-structural protein; SARS-CoV-2, COVID-19, genome-wide, evolution, variants, VOC, VOI, VUM, SNP, mutation, non-synonymous, silent mutation, spike, RNA dependent RNA polymerase, NSP, UTR: Abbreviations: VOC, variant of concern; UTR, untranslated region; VOI, variant of interest; VUM, variant under monitoring; rdrp, RNA dependent RNA polymerase.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The author declares no competing interest.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. - PubMed
-
- Bansal K., Kumar S., Patil P.B. bioRxiv; 2021. Codon usage pattern reveals SARS-CoV-2 as a monomorphic pathogen of hybrid origin with role of silent mutations in rapid evolutionary success. 2020.2010.2012.335521. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

