The Nrf1 transcription factor is induced by patulin and protects against patulin cytotoxicity
- PMID: 35367319
- PMCID: PMC9522914
- DOI: 10.1016/j.tox.2022.153173
The Nrf1 transcription factor is induced by patulin and protects against patulin cytotoxicity
Abstract
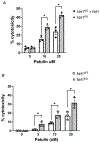
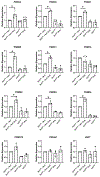
Patulin is a mycotoxin produced by a variety of molds that is found in various food products. The adverse health effects associated with exposure to patulin has led to many investigations into the biological basis driving the toxicity of patulin. Nevertheless, the mechanisms through which mammalian cells resists patulin-mediated toxicity is poorly understood. Here, we show that loss of the Nrf1 transcription factor renders cells sensitive to the acute cytotoxic effects of patulin. Nrf1 deficiency leads to accumulation of ubiquitinated proteins and protein aggregates in response to patulin exposure. Nrf1 expression is induced by patulin, and activation of proteasome genes by patulin is Nrf1-dependent. These findings suggest the Nrf1 transcription factor plays a crucial role in modulating cellular stress response against patulin cytotoxicity.
Keywords: Cellular stress; Mycotoxin; Nrf1; Protein homeostasis; Transcription factor.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures