Probing the mutation independent interaction of DNA probes with SARS-CoV-2 variants through a combination of surface-enhanced Raman scattering and machine learning
- PMID: 35367703
- PMCID: PMC8938299
- DOI: 10.1016/j.bios.2022.114200
Probing the mutation independent interaction of DNA probes with SARS-CoV-2 variants through a combination of surface-enhanced Raman scattering and machine learning
Abstract
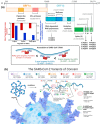
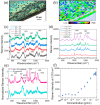
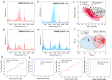
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) evolution has been characterized by the emergence of sets of mutations impacting the virus characteristics, such as transmissibility and antigenicity, presumably in response to the changing immune profile of the human population. The presence of mutations in the SARS-CoV-2 virus can potentially impact therapeutic and diagnostic test performances. We design and develop here a unique set of DNA probes i.e., antisense oligonucleotides (ASOs) which can interact with genetic sequences of the virus irrespective of its ongoing mutations. The probes, developed herein, target a specific segment of the nucleocapsid phosphoprotein (N) gene of SARS-CoV-2 with high binding efficiency which do not mutate among the known variants. Further probing into the interaction profile of the ASOs reveals that the ASO-RNA hybridization remains unaltered even for a hypothetical single point mutation at the target RNA site and diminished only in case of the hypothetical double or triple point mutations. The mechanism of interaction among the ASOs and SARS-CoV-2 RNA is then explored with a combination of surface-enhanced Raman scattering (SERS) and machine learning techniques. It has been observed that the technique, described herein, could efficiently discriminate between clinically positive and negative samples with ∼100% sensitivity and ∼90% specificity up to 63 copies/mL of SARS-CoV-2 RNA concentration. Thus, this study establishes N gene targeted ASOs as the fundamental machinery to efficiently detect all the current SARS-CoV-2 variants regardless of their mutations.
Keywords: Antisense oligonucleotide; Mutation resistant probe; SARS-CoV-2 variants; Selective and ultrasensitive diagnosis; Surface-enhanced Raman scattering.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
D.P. is the founder or co-founder for four University based start ups. None of these entities, however, supported this work.
Figures



References
-
- Abid Hasan S.M., He Y., Chang T.-W., Wang J., Gartia M.R. Detecting DNA methylation using surface-enhanced Raman spectroscopy. J. Phys. Chem. C. 2019;123:698–709. doi: 10.1021/acs.jpcc.8b10178. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

