SARS-CoV-2 in Spent Dialysate from Chronic Peritoneal Dialysis Patients with COVID-19
- PMID: 35368814
- PMCID: PMC8785743
- DOI: 10.34067/KID.0006102020
SARS-CoV-2 in Spent Dialysate from Chronic Peritoneal Dialysis Patients with COVID-19
Abstract
Background: To date, it is unclear whether SARS-CoV-2 is present in spent dialysate from patients with COVID-19 on peritoneal dialysis (PD). Our aim was to assess the presence or absence of SARS-CoV-2 in spent dialysate from patients on chronic PD who had a confirmed diagnosis of COVID-19.
Methods: Spent PD dialysate samples from patients on PD who were positive for COVID-19 were collected between March and August 2020. The multiplexed, real-time RT-PCR assay contained primer/probe sets specific to different SARS-CoV-2 genomic regions and to bacteriophage MS2 as an internal process control for nucleic acid extraction. Demographic and clinical data were obtained from patients' electronic health records.
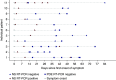
Results: A total of 26 spent PD dialysate samples were collected from 11 patients from ten dialysis centers. Spent PD dialysate samples were collected, on average, 25±13 days (median, 20; range, 10-45) after the onset of symptoms. The temporal distance of PD effluent collection relative to the closest positive nasal-swab RT-PCR result was 15±11 days (median, 14; range, 1-41). All 26 PD effluent samples tested negative at three SARS-CoV-2 genomic regions.
Conclusions: Our findings indicate the absence of SARS-CoV-2 in spent PD dialysate collected at ≥10 days after the onset of COVID-19 symptoms. We cannot rule out the presence of SARS-CoV-2 in spent PD dialysate in the early stage of COVID-19.
Keywords: COVID-19; RT-PCR; SARS-CoV-2; chronic kidney disease; dialysis; dialysis solutions; peritoneal dialysis.
Copyright © 2021 by the American Society of Nephrology.
Conflict of interest statement
J.E. Chao, G.F. Dias, N. Grobe, M. Hakim, M. Han, O. Thwin, Z. Haq, P. Kotanko, A. Patel, P. Preciado, J. Raimann, X. Tao, L. Tisdale, X. Wang, and X. Ye are employees of the Renal Research Institute, a wholly owned subsidiary of FMCNA. D. Chatoth and R. Lasky are employees of FMCNA. D. Chatoth and P. Kotanko hold stock in Fresenius Medical Care. All remaining authors have nothing to disclose.
Figures

References
-
- Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, Men D, Huang Q, Liu Y, Yang B, Ding J, Li F: Detectable serum severe acute respiratory syndrome-coronavirus 2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus 2019. Clin Infect Dis 71: 1937–1942, 2020 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

