Appraisal of Cinnamaldehyde Analogs as Dual-Acting Antibiofilm and Anthelmintic Agents
- PMID: 35369516
- PMCID: PMC8966877
- DOI: 10.3389/fmicb.2022.818165
Appraisal of Cinnamaldehyde Analogs as Dual-Acting Antibiofilm and Anthelmintic Agents
Abstract
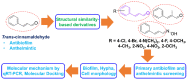
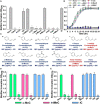
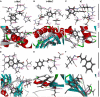
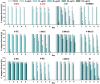
Cinnamaldehyde has a broad range of biological activities, which include antibiofilm and anthelmintic activities. The ever-growing problem of drug resistance and limited treatment options have created an urgent demand for natural molecules with antibiofilm and anthelmintic properties. Hence, we hypothesized that molecules with a scaffold structurally similar to that of cinnamaldehyde might act as dual inhibitors against fungal biofilms and helminths. In this regard, eleven cinnamaldehyde analogs were tested to determine their effects on fungal Candida albicans biofilm and nematode Caenorhabditis elegans. α-Methyl and trans-4-methyl cinnamaldehydes efficiently inhibited C. albicans biofilm formation (>90% inhibition at 50 μg/mL) with minimum inhibitory concentrations (MICs) of ≥ 200 μg/mL and 4-bromo and 4-chloro cinnamaldehydes exhibited anthelmintic property at 20 μg/mL against C. elegans. α-Methyl and trans-4-methyl cinnamaldehydes inhibited hyphal growth and cell aggregation. Scanning electron microscopy was employed to determine the surface architecture of C. albicans biofilm and cuticle of C. elegans, and confocal laser scanning microscopy was used to determine biofilm characteristics. The perturbation in gene expression of C. albicans was investigated using qRT-PCR analysis and α-methyl and trans-4-methyl cinnamaldehydes exhibited down-regulation of ECE1, IFD6, RBT5, UCF1, and UME6 and up-regulation of CHT4 and YWP1. Additionally, molecular interaction of these two molecules with UCF1 and YWP1 were revealed by molecular docking simulation. Our observations collectively suggest α-methyl and trans-4-methyl cinnamaldehydes are potent biofilm inhibitors and that 4-bromo and 4-chloro cinnamaldehydes are anthelmintic agents. Efforts are required to determine the range of potential therapeutic applications of cinnamaldehyde analogs.
Keywords: Candida albicans; anthelmintic; antibiofilm; protein interaction; trans-4-methyl cinnamaldehyde; α-methyl cinnamaldehyde.
Copyright © 2022 Khadke, Lee, Kim, Raj and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Antifungal and antibiofilm activities of flavonoids against Candida albicans: Focus on 3,2'-dihydroxyflavone as a potential therapeutic agent.Biofilm. 2024 Jul 26;8:100218. doi: 10.1016/j.bioflm.2024.100218. eCollection 2024 Dec. Biofilm. 2024. PMID: 39175909 Free PMC article.
-
Antibiofilm activity of lawsone against polymicrobial enterohemorrhagic Escherichia coli O157:H7 and Candida albicans by suppression of curli production and hyphal growth.Phytomedicine. 2024 Feb;124:155306. doi: 10.1016/j.phymed.2023.155306. Epub 2023 Dec 19. Phytomedicine. 2024. PMID: 38176270
-
Biofilm and hyphal inhibitory synergistic effects of phytoactives piperine and cinnamaldehyde against Candida albicans.Med Mycol. 2022 Aug 10;60(8):myac039. doi: 10.1093/mmy/myac039. Med Mycol. 2022. PMID: 35661216
-
Alizarin and Chrysazin Inhibit Biofilm and Hyphal Formation by Candida albicans.Front Cell Infect Microbiol. 2017 Oct 16;7:447. doi: 10.3389/fcimb.2017.00447. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29085811 Free PMC article.
-
Direct one-pot synthesis of cinnamaldehyde immobilized on gold nanoparticles and their antibiofilm properties.Colloids Surf B Biointerfaces. 2017 Dec 1;160:639-648. doi: 10.1016/j.colsurfb.2017.10.018. Epub 2017 Oct 6. Colloids Surf B Biointerfaces. 2017. PMID: 29031224
Cited by
-
Antibiofilm Activities of Cinnamaldehyde Analogs against Uropathogenic Escherichia coli and Staphylococcus aureus.Int J Mol Sci. 2022 Jun 29;23(13):7225. doi: 10.3390/ijms23137225. Int J Mol Sci. 2022. PMID: 35806244 Free PMC article.
-
Unraveling anthelmintic targets and mechanisms of action of trans-cinnamaldehyde from cinnamon essential oil.Sci Rep. 2025 Feb 13;15(1):5422. doi: 10.1038/s41598-025-89883-4. Sci Rep. 2025. PMID: 39948358 Free PMC article.
-
Bioinspired chitosan based functionalization of biomedical implant surfaces for enhanced hemocompatibility, antioxidation and anticoagulation potential: an in silico and in vitro study.RSC Adv. 2024 Jul 1;14(29):20691-20713. doi: 10.1039/d4ra00796d. eCollection 2024 Jun 27. RSC Adv. 2024. PMID: 38952927 Free PMC article.
References
-
- Banerjee M., Uppuluri P., Zhao X. R., Carlisle P. L., Vipulanandan G., Villar C. C., et al. (2013). Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12 224–232. 10.1128/EC.00163-12 - DOI - PMC - PubMed
-
- Beema Shafreen R. M., Selvaraj C., Singh S. K., Karutha Pandian S. (2014). In silico and in vitro studies of cinnamaldehyde and their derivatives against LuxS in Streptococcus pyogenes: effects on biofilm and virulence genes. J. Mol. Recognit. 27 106–116. - PubMed
LinkOut - more resources
Full Text Sources

