A New Way to Trace SARS-CoV-2 Variants Through Weighted Network Analysis of Frequency Trajectories of Mutations
- PMID: 35369526
- PMCID: PMC8966897
- DOI: 10.3389/fmicb.2022.859241
A New Way to Trace SARS-CoV-2 Variants Through Weighted Network Analysis of Frequency Trajectories of Mutations
Abstract
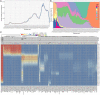
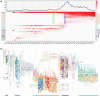
Early detection of SARS-CoV-2 variants enables timely tracking of clinically important strains in order to inform the public health response. Current subtype-based variant surveillance depending on prior subtype assignment according to lag features and their continuous risk assessment may delay this process. We proposed a weighted network framework to model the frequency trajectories of mutations (FTMs) for SARS-CoV-2 variant tracing, without requiring prior subtype assignment. This framework modularizes the FTMs and conglomerates synchronous FTMs together to represent the variants. It also generates module clusters to unveil the epidemic stages and their contemporaneous variants. Eventually, the module-based variants are assessed by phylogenetic tree through sub-sampling to facilitate communication and control of the epidemic. This process was benchmarked using worldwide GISAID data, which not only demonstrated all the methodology features but also showed the module-based variant identification had highly specific and sensitive mapping with the global phylogenetic tree. When applying this process to regional data like India and South Africa for SARS-CoV-2 variant surveillance, the approach clearly elucidated the national dispersal history of the viral variants and their co-circulation pattern, and provided much earlier warning of Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529). In summary, our work showed that the weighted network modeling of FTMs enables us to rapidly and easily track down SARS-CoV-2 variants overcoming prior viral subtyping with lag features, accelerating the understanding and surveillance of COVID-19.
Keywords: SARS-CoV-2; frequency trajectories; mutations; variant tracing; weighted network analysis.
Copyright © 2022 Huang, Zhang, Bible, Liang, Zheng, Wang, Hao and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Csárdi G., Nepusz T. (2006). The igraph software package for complex network research. InterJ. Complex Syst. 1695 1–9.
LinkOut - more resources
Full Text Sources
Miscellaneous

