A Comprehensive Network Integrating Signature Microbes and Crucial Soil Properties During Early Biological Soil Crust Formation on Tropical Reef Islands
- PMID: 35369528
- PMCID: PMC8969229
- DOI: 10.3389/fmicb.2022.831710
A Comprehensive Network Integrating Signature Microbes and Crucial Soil Properties During Early Biological Soil Crust Formation on Tropical Reef Islands
Abstract
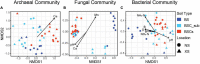
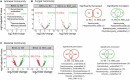
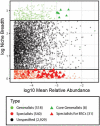
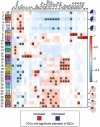
Biological soil crusts (BSCs/biocrusts), which are distributed across various climatic zones and well-studied in terrestrial drylands, harbor polyextremotolerant microbial topsoil communities and provide ecological service for local and global ecosystem. Here, we evaluated BSCs in the tropical reef islands of the South China Sea. Specifically, we collected 41 BSCs, subsurface, and bare soil samples from the Xisha and Nansha Archipelagos. High-throughput amplicon sequencing was performed to analyze the bacterial, fungal, and archaeal compositions of these samples. Physicochemical measurement and enzyme activity assays were conducted to characterize the soil properties. Advanced computational analysis revealed 47 biocrust-specific microbes and 10 biocrust-specific soil properties, as well as their correlations in BSC microbial community. We highlighted the previously underestimated impact of manganese on fungal community regulation and BSC formation. We provide comprehensive insight into BSC formation networks on tropical reef islands and established a foundation for BSC-directed environmental restoration.
Keywords: biocrust formation; biological soil crust; enzyme activity; geographical distribution pattern; microbiome; tropical reef island.
Copyright © 2022 Wang, Li and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aanderud Z. T., Smart T. B., Wu N., Taylor A. S., Zhang Y., Belnap J. (2018). Fungal loop transfer of nitrogen depends on biocrust constituents and nitrogen form. Biogeosciences 15 3831–3840. 10.5194/bg-15-3831-2018 - DOI
-
- Acosta-Martínez V., Cruz L., Sotomayor-Ramírez D., Pérez-Alegría L. (2007). Enzyme activities as affected by soil properties and land use in a tropical watershed. Appl. Soil Ecol. 35 35–45. 10.1016/j.apsoil.2006.05.012 - DOI
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26 32–46. 10.1111/j.1442-9993.2001.01070.pp.x - DOI
LinkOut - more resources
Full Text Sources

