Multi-omics analysis identifies distinct subtypes with clinical relevance in lung adenocarcinoma harboring KEAP1/ NFE2L2
- PMID: 35371318
- PMCID: PMC8965119
- DOI: 10.7150/jca.66241
Multi-omics analysis identifies distinct subtypes with clinical relevance in lung adenocarcinoma harboring KEAP1/ NFE2L2
Abstract
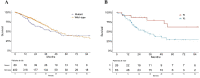
Backgrounds: Lung adenocarcinoma is one of the most common malignant tumors, in which KEAP1-NFE2L2 pathway is altered frequently. The biological features and intrinsic heterogeneities of KEAP1/NFE2L2-mutant lung adenocarcinoma remain unclear. Methods: Multiplatform data from The Cancer Genome Atlas (TCGA) were acquired to identify two subtypes of lung adenocarcinoma harboring KEAP1/NFE2L2 mutations. Bioinformatic analyses, including immune microenvironment, methylation level and mutational signature, were performed to characterize the intrinsic heterogeneities. Meanwhile, initial results were validated by using in silico assessment of common lung adenocarcinoma cell lines, which revealed consistent features of mutant subtypes. Furthermore, drug sensitivity screening was conducted based on public datasets. Results: Two mutant subtypes (P1 and P2) of 89 patients were identified in TCGA. P2 patients had significantly higher levels of smoking and worse survival compared with P1 patients. The P2 subset was characterized by active immune microenvironment and more smoking-induced genomic alterations with respect to methylation and somatic mutations. Validations of the corresponding features in 20 mutant cell lines were achieved. Several compounds which were sensitive to mutant subtypes of lung adenocarcinoma were identified, such as inhibitors of PI3K/Akt and IGF1R signaling pathways. Conclusions: KEAP1/NFE2L2-mutant lung adenocarcinoma showed potential heterogeneities. The intrinsic heterogeneities of KEAP1/NFE2L2 were associated with immune microenvironment and smoking-related genomic aberrations.
Keywords: KEAP1; NFE2L2; lung adenocarcinoma; mutation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





Similar articles
-
Mutations in the KEAP1-NFE2L2 Pathway Define a Molecular Subset of Rapidly Progressing Lung Adenocarcinoma.J Thorac Oncol. 2019 Nov;14(11):1924-1934. doi: 10.1016/j.jtho.2019.07.003. Epub 2019 Jul 16. J Thorac Oncol. 2019. PMID: 31323387
-
Integrated analysis of patients with KEAP1/NFE2L2/CUL3 mutations in lung adenocarcinomas.Cancer Med. 2021 Dec;10(23):8673-8692. doi: 10.1002/cam4.4338. Epub 2021 Oct 6. Cancer Med. 2021. PMID: 34617407 Free PMC article.
-
Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2-mutant lung cancer.Int J Cancer. 2019 Feb 15;144(4):788-801. doi: 10.1002/ijc.31975. Epub 2018 Dec 4. Int J Cancer. 2019. PMID: 30411339
-
Targeting the cell signaling pathway Keap1-Nrf2 as a therapeutic strategy for adenocarcinomas of the lung.Expert Opin Ther Targets. 2019 Mar;23(3):241-250. doi: 10.1080/14728222.2019.1559824. Epub 2018 Dec 27. Expert Opin Ther Targets. 2019. PMID: 30556750 Review.
-
KEAP1/NRF2 (NFE2L2) mutations in NSCLC - Fuel for a superresistant phenotype?Lung Cancer. 2021 Sep;159:10-17. doi: 10.1016/j.lungcan.2021.07.006. Epub 2021 Jul 17. Lung Cancer. 2021. PMID: 34303275 Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
LinkOut - more resources
Full Text Sources

