First Report of a Methicillin-Resistant, High-Level Mupirocin-Resistant Staphylococcus argenteus
- PMID: 35372120
- PMCID: PMC8964999
- DOI: 10.3389/fcimb.2022.860163
First Report of a Methicillin-Resistant, High-Level Mupirocin-Resistant Staphylococcus argenteus
Abstract
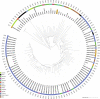
We describe the identification of a methicillin-resistant, high-level mupirocin-resistant Staphylococcus argenteus. The isolate (1801221) was characterized as t6675-ST2250-SCCmecIVc, and whole-genome sequencing revealed that the isolate possessed two plasmids. One plasmid (34,870 bp), designated p1_1801221 with rep23, harboured the mupirocin resistance (mupA) gene. The second plasmid (20,644 bp), assigned as p2_1801221 with rep5a and rep16, carried the resistance determinants for penicillin (blaZ) and cadmium (cadD). Phylogenetic analysis revealed that the isolate clustered with the European ST2250 lineage. The overall high similarity of both plasmids in S. argenteus with published DNA sequences of Staphylococcus aureus plasmids strongly suggests an interspecies transfer. The pathogenic potential, community and nosocomial spread, and acquisition of antibiotic resistance gene determinants, including the mupA gene by S. argenteus, highlight its clinical significance and the need for its correct identification.
Keywords: high-level mupirocin resistance; identification; methicillin-resistant Staphylococcus argenteus; plasmid; whole-genome sequencing (WGS).
Copyright © 2022 Shittu, Layer-Nicolaou, Strommenger, Nguyen, Bletz, Mellmann and Schaumburg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Alhussein F., Fürstenberg J., Gaupp R., Eisenbeis J., Last K., Becker S. L., et al. . (2020). Human Infections Caused by Staphylococcus Argenteus in Germany: Genetic Characterisation and Clinical Implications of Novel Species Designation. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc Clin. Microbiol. 39, 2461–2465. doi: 10.1007/s10096-020-03950-4 - DOI - PMC - PubMed
-
- Aung M. S., San T., Aye M. M., Mya S., Maw W. W., Zan K. N., et al. . (2017). Prevalence and Genetic Characteristics of Staphylococcus Aureus and Staphylococcus Argenteus Isolates Harboring Panton-Valentine Leukocidin, Enterotoxins, and TSST-1 Genes From Food Handlers in Myanmar. Toxins (Basel) 9 (241), 1–13. doi: 10.3390/toxins9080241 - DOI - PMC - PubMed
-
- Aung M. S., Urushibara N., Kawaguchiya M., Hirose M., Ike M., Ito M., et al. . (2021). Distribution of Virulence Factors and Resistance Determinants in Three Genotypes of Staphylococcus Argenteus Clinical Isolates in Japan. Pathog. (Basel Switzerland) 10, 163. doi: 10.3390/pathogens10020163 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

