An explainable artificial intelligence approach for predicting cardiovascular outcomes using electronic health records
- PMID: 35373216
- PMCID: PMC8975108
- DOI: 10.1371/journal.pdig.0000004
An explainable artificial intelligence approach for predicting cardiovascular outcomes using electronic health records
Abstract
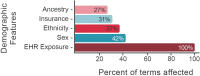
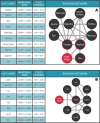
Understanding the conditionally-dependent clinical variables that drive cardiovascular health outcomes is a major challenge for precision medicine. Here, we deploy a recently developed massively scalable comorbidity discovery method called Poisson Binomial based Comorbidity discovery (PBC), to analyze Electronic Health Records (EHRs) from the University of Utah and Primary Children's Hospital (over 1.6 million patients and 77 million visits) for comorbid diagnoses, procedures, and medications. Using explainable Artificial Intelligence (AI) methodologies, we then tease apart the intertwined, conditionally-dependent impacts of comorbid conditions and demography upon cardiovascular health, focusing on the key areas of heart transplant, sinoatrial node dysfunction and various forms of congenital heart disease. The resulting multimorbidity networks make possible wide-ranging explorations of the comorbid and demographic landscapes surrounding these cardiovascular outcomes, and can be distributed as web-based tools for further community-based outcomes research. The ability to transform enormous collections of EHRs into compact, portable tools devoid of Protected Health Information solves many of the legal, technological, and data-scientific challenges associated with large-scale EHR analyses.
Conflict of interest statement
Competing interests: I have read the journal’s policy and the authors of this manuscript have the following competing interests: GL, VD, MY own shares in Backdrop Health, there are no financial ties regarding this research.
Figures



References
-
- Kraisangka J. et al.. Bayesian Network vs. Cox’s Proportional Hazard Model of PAH Risk: A Comparison. in Artificial Intelligence in Medicine (eds. Riaño D., Wilk S. & ten Teije A.) 139–149 (Springer International Publishing, 2019). doi: 10.1007/s11906-019-0950-y - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
