Identification and validation of a ferroptosis-related gene signature for predicting survival in skin cutaneous melanoma
- PMID: 35373463
- PMCID: PMC9487883
- DOI: 10.1002/cam4.4706
Identification and validation of a ferroptosis-related gene signature for predicting survival in skin cutaneous melanoma
Abstract
Purpose: Ferroptosis plays a crucial role in the initiation and progression of melanoma. This study developed a robust signature with ferroptosis-related genes (FRGs) and assessed the ability of this signature to predict OS in patients with skin cutaneous melanoma (SKCM).
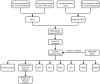
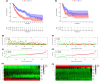
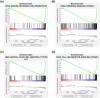
Methods: RNA-sequencing data and clinical information of melanoma patients were extracted from TCGA, GEO, and GTEx. Univariate, multivariate, and LASSO regression analyses were conducted to identify the gene signature. A 10 FRG signature was an independent and strong predictor of survival. The predictive performance was assessed using ROC curve. The functions of this gene signature were assessed by GO and KEGG analysis. The statuses of low-risk and high-risk groups according to the gene signature were compared by GSEA. In addition, we investigated the possible relationship of FRGs with immunotherapy efficacy.
Results: A prognostic signature with 10 FRGs (CYBB, IFNG, FBXW7, ARNTL, PROM2, GPX2, JDP2, SLC7A5, TUBE1, and HAMP) was identified by Cox regression analysis. This signature had a higher prediction efficiency than clinicopathological features (AUC = 0.70). The enrichment analyses of DEGs indicated that ferroptosis-related immune pathways were largely enriched. Furthermore, GSEA showed that ferroptosis was associated with immunosuppression in the high-risk group. Finally, immune checkpoints such as PDCD-1 (PD-1), CTLA4, CD274 (PD-L1), and LAG3 were also differential expression in two risk groups.
Conclusions: The 10 FRGs signature were a strong predictor of OS in SKCM and could be used to predict therapeutic targets for melanoma.
Keywords: ferroptosis; gene signature; melanoma; overall survival; tumor immunity.
© 2022 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest in this work.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

