Bottom-Up Assembly of Synthetic Cells with a DNA Cytoskeleton
- PMID: 35377150
- PMCID: PMC9134502
- DOI: 10.1021/acsnano.1c10703
Bottom-Up Assembly of Synthetic Cells with a DNA Cytoskeleton
Abstract
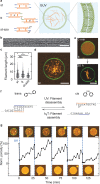
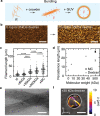
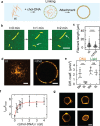
Cytoskeletal elements, like actin and myosin, have been reconstituted inside lipid vesicles toward the vision to reconstruct cells from the bottom up. Here, we realize the de novo assembly of entirely artificial DNA-based cytoskeletons with programmed multifunctionality inside synthetic cells. Giant unilamellar lipid vesicles (GUVs) serve as cell-like compartments, in which the DNA cytoskeletons are repeatedly and reversibly assembled and disassembled with light using the cis-trans isomerization of an azobenzene moiety positioned in the DNA tiles. Importantly, we induced ordered bundling of hundreds of DNA filaments into more rigid structures with molecular crowders. We quantify and tune the persistence length of the bundled filaments to achieve the formation of ring-like cortical structures inside GUVs, resembling actin rings that form during cell division. Additionally, we show that DNA filaments can be programmably linked to the compartment periphery using cholesterol-tagged DNA as a linker. The linker concentration determines the degree of the cortex-like network formation, and we demonstrate that the DNA cortex-like network can deform GUVs from within. All in all, this showcases the potential of DNA nanotechnology to mimic the diverse functions of a cytoskeleton in synthetic cells.
Keywords: DNA nanotechnology; DNA nanotube; azobenzene; bottom-up synthetic biology; giant unilamellar vesicles; synthetic cell.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Bashirzadeh Y.; Redford S. A.; Lorpaiboon C.; Groaz A.; Moghimianavval H.; Litschel T.; Schwille P.; Hocky G. M.; Dinner A. R.; Liu A. P. Actin Crosslinker Competition and Sorting Drive Emergent GUV Size-Dependent Actin Network Architecture. Communications Biology 2021, 4, 1136. 10.1038/s42003-021-02653-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

