The Ahr2-Dependent wfikkn1 Gene Influences Zebrafish Transcriptome, Proteome, and Behavior
- PMID: 35377459
- PMCID: PMC9308396
- DOI: 10.1093/toxsci/kfac037
The Ahr2-Dependent wfikkn1 Gene Influences Zebrafish Transcriptome, Proteome, and Behavior
Abstract
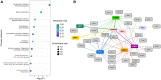
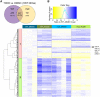
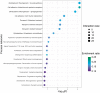
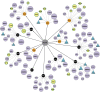
The aryl hydrocarbon receptor (AHR) is required for vertebrate development and is also activated by exogenous chemicals, including polycyclic aromatic hydrocarbons (PAHs) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). AHR activation is well-understood, but roles of downstream molecular signaling events are largely unknown. From previous transcriptomics in 48 h postfertilization (hpf) zebrafish exposed to several PAHs and TCDD, we found wfikkn1 was highly coexpressed with cyp1a (marker for AHR activation). Thus, we hypothesized wfikkn1's role in AHR signaling, and showed that wfikkn1 expression was Ahr2 (zebrafish ortholog of human AHR)-dependent in developing zebrafish exposed to TCDD. To functionally characterize wfikkn1, we made a CRISPR-Cas9 mutant line with a 16-bp deletion in wfikkn1's exon, and exposed wildtype and mutants to dimethyl sulfoxide or TCDD. 48-hpf mRNA sequencing revealed over 700 genes that were differentially expressed (p < .05, log2FC > 1) between each pair of treatment combinations, suggesting an important role for wfikkn1 in altering both the 48-hpf transcriptome and TCDD-induced expression changes. Mass spectrometry-based proteomics of 48-hpf wildtype and mutants revealed 325 significant differentially expressed proteins. Functional enrichment demonstrated wfikkn1 was involved in skeletal muscle development and played a role in neurological pathways after TCDD exposure. Mutant zebrafish appeared morphologically normal but had significant behavior deficiencies at all life stages, and absence of Wfikkn1 did not significantly alter TCDD-induced behavior effects at all life stages. In conclusion, wfikkn1 did not appear to be significantly involved in TCDD's overt toxicity but is likely a necessary functional member of the AHR signaling cascade.
Keywords: TCDD; aryl hydrocarbon receptor (AHR); behavior; transcriptomics; wfikkn1; zebrafish.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society of Toxicology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures






References
-
- Ahmad F., Noldus L. P. J. J., Tegelenbosch R. A. J., Richardson M. K. (2012). Zebrafish embryos and larvae in behavioural assays. Behaviour 149, 1241–1281.
-
- Andreasen E. A., Spitsbergen J. M., Tanguay R. L., Stegeman J. J., Heideman W., Peterson R. E. (2002). Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: Effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol. Sci. 68, 403–419. - PubMed
-
- Babin P. J., Goizet C., Raldua D. (2014). Zebrafish models of human motor neuron diseases: Advantages and limitations. Prog. Neurobiol. 118, 36–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

