A generalist-specialist trade-off between switchgrass cytotypes impacts climate adaptation and geographic range
- PMID: 35377798
- PMCID: PMC9169841
- DOI: 10.1073/pnas.2118879119
A generalist-specialist trade-off between switchgrass cytotypes impacts climate adaptation and geographic range
Abstract
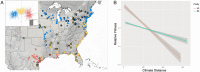
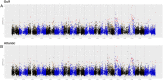
Polyploidy results from whole-genome duplication and is a unique form of heritable variation with pronounced evolutionary implications. Different ploidy levels, or cytotypes, can exist within a single species, and such systems provide an opportunity to assess how ploidy variation alters phenotypic novelty, adaptability, and fitness, which can, in turn, drive the development of unique ecological niches that promote the coexistence of multiple cytotypes. Switchgrass, Panicum virgatum, is a widespread, perennial C4 grass in North America with multiple naturally occurring cytotypes, primarily tetraploids (4×) and octoploids (8×). Using a combination of genomic, quantitative genetic, landscape, and niche modeling approaches, we detect divergent levels of genetic admixture, evidence of niche differentiation, and differential environmental sensitivity between switchgrass cytotypes. Taken together, these findings support a generalist (8×)–specialist (4×) trade-off. Our results indicate that the 8× represent a unique combination of genetic variation that has allowed the expansion of switchgrass’ ecological niche and thus putatively represents a valuable breeding resource.
Keywords: Panicum virgatum; cytotypes; octoploid.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Ramsey J., Schemske D. W., Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501 (1998).
-
- Soltis P. S., Marchant D. B., Van de Peer Y., Soltis D. E., Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125 (2015). - PubMed
-
- Kolář F., Čertner M., Suda J., Schönswetter P., Husband B. C., Mixed-ploidy species: Progress and opportunities in polyploid research. Trends Plant Sci. 22, 1041–1055 (2017). - PubMed
-
- Otto S. P., The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

