NK cells and monocytes modulate primary HTLV-1 infection
- PMID: 35377924
- PMCID: PMC9022856
- DOI: 10.1371/journal.ppat.1010416
NK cells and monocytes modulate primary HTLV-1 infection
Abstract
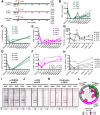
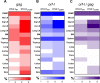
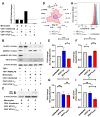
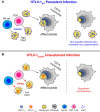
We investigated the impact of monocytes, NK cells, and CD8+ T-cells in primary HTLV-1 infection by depleting cell subsets and exposing macaques to either HTLV-1 wild type (HTLV-1WT) or to the HTLV-1p12KO mutant unable to infect replete animals due to a single point mutation in orf-I that inhibits its expression. The orf-I encoded p8/p12 proteins counteract cytotoxic NK and CD8+ T-cells and favor viral DNA persistence in monocytes. Double NK and CD8+ T-cells or CD8 depletion alone accelerated seroconversion in all animals exposed to HTLV-1WT. In contrast, HTLV-1p12KO infectivity was fully restored only when NK cells were also depleted, demonstrating a critical role of NK cells in primary infection. Monocyte/macrophage depletion resulted in accelerated seroconversion in all animals exposed to HTLV-1WT, but antibody titers to the virus were low and not sustained. Seroconversion did not occur in most animals exposed to HTLV-1p12KO. In vitro experiments in human primary monocytes or THP-1 cells comparing HTLV-1WT and HTLV-1p12KO demonstrated that orf-I expression is associated with inhibition of inflammasome activation in primary cells, with increased CD47 "don't-eat-me" signal surface expression in virus infected cells and decreased monocyte engulfment of infected cells. Collectively, our data demonstrate a critical role for innate NK cells in primary infection and suggest a dual role of monocytes in primary infection. On one hand, orf-I expression increases the chances of viral transmission by sparing infected cells from efferocytosis, and on the other may protect the engulfed infected cells by modulating inflammasome activation. These data also suggest that, once infection is established, the stoichiometry of orf-I expression may contribute to the chronic inflammation observed in HTLV-1 infection by modulating monocyte efferocytosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

