DiffBond: A Method for Predicting Intermolecular Bond Formation
- PMID: 35378834
- PMCID: PMC8976940
- DOI: 10.1109/bibm52615.2021.9669850
DiffBond: A Method for Predicting Intermolecular Bond Formation
Abstract
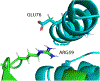
Many tools that explore models of protein complexes are also able to analyze interactions between specific residues and atoms. A comprehensive exploration of these interactions can often uncover aspects of protein-protein recognition that are not obvious using other protein analysis techniques. This paper describes DiffBond, a novel method for searching for intermolecular interactions between protein complexes while differentiating between three different types of interaction: hydrogen bonds, ionic bonds, and salt bridges. DiffBond incorporates textbook definitions of these three interactions while contending with uncertainties that are inherent in computational models of interacting proteins. We used it to examine the barnase-barstar, Rap1a-raf, and Smad2-Smad4 complexes, as well as a subset of protein complexes formed between three-finger toxins and nAChRs. Based on electrostatic interactions established by previous experimental studies, DiffBond was able to identify ionic and hydrogen bonds with high precision and recall, and identify salt bridges with high precision. In combination with other electrostatic analysis methods, DiffBond can be a useful tool in helping predict influential amino acids in protein-protein interactions and characterizing the type of interaction.
Figures

References
-
- McDonald IK and Thornton JM, “Satisfying hydrogen bonding potential in proteins,” Journal of molecular biology, vol. 238, no. 5, pp. 777–793, 1994. - PubMed
-
- Barlow DJ and Thornton J, “Ion-pairs in proteins,” Journal of molecular biology, vol. 168, no. 4, pp. 867–885, 1983. - PubMed
-
- Klapper I, Hagstrom R, Fine R, Sharp K, and Honig B, “Focusing of electric fields in the active site of cu-zn superoxide dismutase: Effects of ionic strength and amino-acid modification,” Proteins: Structure, Function, and Bioinformatics, vol. 1, no. 1, pp. 47–59, 1986. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
