Predicted coronavirus Nsp5 protease cleavage sites in the human proteome
- PMID: 35379171
- PMCID: PMC8977440
- DOI: 10.1186/s12863-022-01044-y
Predicted coronavirus Nsp5 protease cleavage sites in the human proteome
Abstract
Background: The coronavirus nonstructural protein 5 (Nsp5) is a cysteine protease required for processing the viral polyprotein and is therefore crucial for viral replication. Nsp5 from several coronaviruses have also been found to cleave host proteins, disrupting molecular pathways involved in innate immunity. Nsp5 from the recently emerged SARS-CoV-2 virus interacts with and can cleave human proteins, which may be relevant to the pathogenesis of COVID-19. Based on the continuing global pandemic, and emerging understanding of coronavirus Nsp5-human protein interactions, we set out to predict what human proteins are cleaved by the coronavirus Nsp5 protease using a bioinformatics approach.
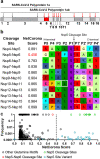
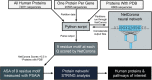
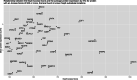
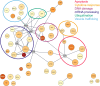
Results: Using a previously developed neural network trained on coronavirus Nsp5 cleavage sites (NetCorona), we made predictions of Nsp5 cleavage sites in all human proteins. Structures of human proteins in the Protein Data Bank containing a predicted Nsp5 cleavage site were then examined, generating a list of 92 human proteins with a highly predicted and accessible cleavage site. Of those, 48 are expected to be found in the same cellular compartment as Nsp5. Analysis of this targeted list of proteins revealed molecular pathways susceptible to Nsp5 cleavage and therefore relevant to coronavirus infection, including pathways involved in mRNA processing, cytokine response, cytoskeleton organization, and apoptosis.
Conclusions: This study combines predictions of Nsp5 cleavage sites in human proteins with protein structure information and protein network analysis. We predicted cleavage sites in proteins recently shown to be cleaved in vitro by SARS-CoV-2 Nsp5, and we discuss how other potentially cleaved proteins may be relevant to coronavirus mediated immune dysregulation. The data presented here will assist in the design of more targeted experiments, to determine the role of coronavirus Nsp5 cleavage of host proteins, which is relevant to understanding the molecular pathology of coronavirus infection.
Keywords: 3CLpro; COVID-19; Coronavirus; Human proteins; Human proteome; Mpro; Nsp5; Protease; SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous
