Efficacy and safety of camrelizumab plus apatinib during the perioperative period in resectable hepatocellular carcinoma: a single-arm, open label, phase II clinical trial
- PMID: 35379737
- PMCID: PMC8981365
- DOI: 10.1136/jitc-2022-004656
Efficacy and safety of camrelizumab plus apatinib during the perioperative period in resectable hepatocellular carcinoma: a single-arm, open label, phase II clinical trial
Abstract
Objective: This study aimed to assess the efficacy and safety of camrelizumab plus apatinib in patients with resectable hepatocellular carcinoma (HCC) as neoadjuvant therapy.
Methods: Initially, 20 patients with HCC were screened and 18 patients with resectable HCC were enrolled in this open-label, single-arm, phase II clinical trial. Patients received three cycles of neoadjuvant therapy including three doses of camrelizumab concurrent with apatinib for 21 days followed by surgery. Four to 8 weeks after surgery, patients received eight cycles of adjuvant therapy with camrelizumab in combination with apatinib. Major pathological reactions (MPR), complete pathological reactions (pCR), objective response rate (ORR), relapse-free survival (RFS), and adverse events (AE) were assessed. In addition, cancer tissue and plasma samples were collected before and after treatment, and genetic differences between responding and non-responding lesions were compared by tumor immune microenvironment (TIME) analysis, circulating tumor DNA (ctDNA) analysis and proteomics analysis.
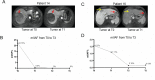
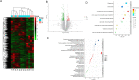
Results: In 18 patients with HCC who completed neoadjuvant therapy, 3 (16.7%) and 6 (33.3%) patients with HCC reached ORR based on Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1 and modified RECIST criteria, respectively. Of the 17 patients with HCC who received surgical resection, 3 (17.6%) patients with HCC reported MPR and 1 (5.9%) patient with HCC achieved pCR. The 1-year RFS rate of the enrolled patients was 53.85% (95% CI: 24.77% to 75.99%). Grade 3/4 AEs were reported in 3 (16.7%) of the 18 patients, with the most common AEs being rash (11.1%), hypertension (5.6%), drug-induced liver damage (5.6%), and neutropenia (5.6%) in the preoperative phase. The 289 NanoString panel RNA sequencing showed that TIME cell infiltration especially dendritic cells (DCs) infiltration was better in responding tumors than in non-responding tumors. Our results of ctDNA revealed a higher positive rate (100%) among patients with HCC with stage IIb-IIIa disease. When comparing patients with pCR/MPR and non-MPR, we observed more mutations in patients who achieved pCR/MPR at baseline (6 mutations vs 2.5 mutations, p=0.025). Patients who were ctDNA positive after adjuvant therapy presented a trend of shorter RFS than those who were ctDNA negative. Proteomic analysis suggested that abnormal glucose metabolism in patients with multifocal HCC might be related to different sensitivity of treatment in different lesions.
Conclusion: Perioperative camrelizumab plus apatinib displays a promising efficacy and manageable toxicity in patients with resectable HCC. DCs infiltration might be a predictive marker of response to camrelizumab and apatinib as well as patients' recurrence. ctDNA as a compose biomarker can predict pathological response and relapse. Abnormal glucose metabolism in patients with multifocal HCC may be related to different sensitivity of treatment in different lesions.
Trial registration number: NCT04297202.
Keywords: antibodies, neoplasm; antineoplastic protocols; immunity; immunotherapy; liver neoplasms.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
