Characterization of Altered Gene Expression and Histone Methylation in Peripheral Blood Mononuclear Cells Regulating Inflammation in COVID-19 Patients
- PMID: 35379747
- PMCID: PMC9012677
- DOI: 10.4049/jimmunol.2101099
Characterization of Altered Gene Expression and Histone Methylation in Peripheral Blood Mononuclear Cells Regulating Inflammation in COVID-19 Patients
Abstract
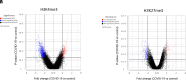
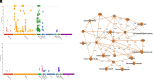
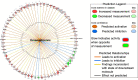
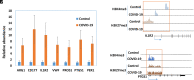
The pandemic of COVID-19 has caused >5 million deaths in the world. One of the leading causes of the severe form of COVID-19 is the production of massive amounts of proinflammatory cytokines. Epigenetic mechanisms, such as histone/DNA methylation, miRNA, and long noncoding RNA, are known to play important roles in the regulation of inflammation. In this study, we investigated if hospitalized COVID-19 patients exhibit alterations in epigenetic pathways in their PBMCs. We also compared gene expression profiles between healthy controls and COVID-19 patients. Despite individual variations, the expressions of many inflammation-related genes, such as arginase 1 and IL-1 receptor 2, were significantly upregulated in COVID-19 patients. We also found the expressions of coagulation-related genes Von Willebrand factor and protein S were altered in COVID-19 patients. The expression patterns of some genes, such as IL-1 receptor 2, correlated with their histone methylation marks. Pathway analysis indicated that most of those dysregulated genes were in the TGF-β, IL-1b, IL-6, and IL-17 pathways. A targeting pathway revealed that the majority of those altered genes were targets of dexamethasone, which is an approved drug for COVID-19 treatment. We also found that the expression of bone marrow kinase on chromosome X, a member of TEC family kinases, was increased in the PBMCs of COVID-19 patients. Interestingly, some inhibitors of TEC family kinases have been used to treat COVID-19. Overall, this study provides important information toward identifying potential biomarkers and therapeutic targets for COVID-19 disease.
Copyright © 2022 by The American Association of Immunologists, Inc.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

