Targeted protein degradation: mechanisms, strategies and application
- PMID: 35379777
- PMCID: PMC8977435
- DOI: 10.1038/s41392-022-00966-4
Targeted protein degradation: mechanisms, strategies and application
Abstract
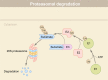
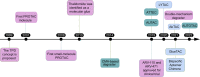
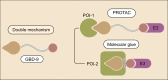
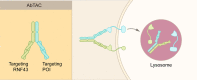
Traditional drug discovery mainly focuses on direct regulation of protein activity. The development and application of protein activity modulators, particularly inhibitors, has been the mainstream in drug development. In recent years, PROteolysis TArgeting Chimeras (PROTAC) technology has emerged as one of the most promising approaches to remove specific disease-associated proteins by exploiting cells' own destruction machinery. In addition to PROTAC, many different targeted protein degradation (TPD) strategies including, but not limited to, molecular glue, Lysosome-Targeting Chimaera (LYTAC), and Antibody-based PROTAC (AbTAC), are emerging. These technologies have not only greatly expanded the scope of TPD, but also provided fresh insights into drug discovery. Here, we summarize recent advances of major TPD technologies, discuss their potential applications, and hope to provide a prime for both biologists and chemists who are interested in this vibrant field.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

