Recovery of mitogenomes from whole genome sequences to infer maternal diversity in 1883 modern taurine and indicine cattle
- PMID: 35379858
- PMCID: PMC8980051
- DOI: 10.1038/s41598-022-09427-y
Recovery of mitogenomes from whole genome sequences to infer maternal diversity in 1883 modern taurine and indicine cattle
Abstract
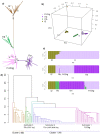
Maternal diversity based on a sub-region of mitochondrial genome or variants were commonly used to understand past demographic events in livestock. Additionally, there is growing evidence of direct association of mitochondrial genetic variants with a range of phenotypes. Therefore, this study used complete bovine mitogenomes from a large sequence database to explore the full spectrum of maternal diversity. Mitogenome diversity was evaluated among 1883 animals representing 156 globally important cattle breeds. Overall, the mitogenomes were diverse: presenting 11 major haplogroups, expanding to 1309 unique haplotypes, with nucleotide diversity 0.011 and haplotype diversity 0.999. A small proportion of African taurine (3.5%) and indicine (1.3%) haplogroups were found among the European taurine breeds and composites. The haplogrouping was largely consistent with the population structure derived from alternate clustering methods (e.g. PCA and hierarchical clustering). Further, we present evidence confirming a new indicine subgroup (I1a, 64 animals) mainly consisting of breeds originating from China and characterised by two private mutations within the I1 haplogroup. The total genetic variation was attributed mainly to within-breed variance (96.9%). The accuracy of the imputation of missing genotypes was high (99.8%) except for the relatively rare heteroplasmic genotypes, suggesting the potential for trait association studies within a breed.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

