Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus
- PMID: 35379995
- PMCID: PMC9664907
- DOI: 10.1038/s41593-022-01043-3
Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus
Abstract
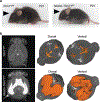
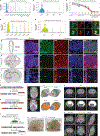
Hydrocephalus, characterized by cerebral ventricular dilatation, is routinely attributed to primary defects in cerebrospinal fluid (CSF) homeostasis. This fosters CSF shunting as the leading reason for brain surgery in children despite considerable disease heterogeneity. In this study, by integrating human brain transcriptomics with whole-exome sequencing of 483 patients with congenital hydrocephalus (CH), we found convergence of CH risk genes in embryonic neuroepithelial stem cells. Of all CH risk genes, TRIM71/lin-41 harbors the most de novo mutations and is most specifically expressed in neuroepithelial cells. Mice harboring neuroepithelial cell-specific Trim71 deletion or CH-specific Trim71 mutation exhibit prenatal hydrocephalus. CH mutations disrupt TRIM71 binding to its RNA targets, causing premature neuroepithelial cell differentiation and reduced neurogenesis. Cortical hypoplasia leads to a hypercompliant cortex and secondary ventricular enlargement without primary defects in CSF circulation. These data highlight the importance of precisely regulated neuroepithelial cell fate for normal brain-CSF biomechanics and support a clinically relevant neuroprogenitor-based paradigm of CH.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
J.J. is an employee of GeneDx, Inc.
Figures
















References
-
- Aristotle. The History of Animals http://classics.mit.edu/Aristotle/history_anim.mb.txt
-
- Persson EK, Hagberg G. & Uvebrant P. Disabilities in children with hydrocephalus—a population-based study of children aged between four and twelve years. Neuropediatrics 37, 330–336 (2006). - PubMed
-
- Greenberg MS Handbook of Neurosurgery, 9th edition (Thieme, 2019).
Publication types
MeSH terms
Substances
Grants and funding
- F30 HD106694/HD/NICHD NIH HHS/United States
- RF1 NS113278/NS/NINDS NIH HHS/United States
- R01 NS111029/NS/NINDS NIH HHS/United States
- R37 DA023999/DA/NIDA NIH HHS/United States
- R01 NS127879/NS/NINDS NIH HHS/United States
- DP2 AI138259/AI/NIAID NIH HHS/United States
- R01 NS035129/NS/NINDS NIH HHS/United States
- R21 NS116484/NS/NINDS NIH HHS/United States
- R01 DA023999/DA/NIDA NIH HHS/United States
- R01 MH113257/MH/NIMH NIH HHS/United States
- T32 GM136651/GM/NIGMS NIH HHS/United States
- P50 HD105351/HD/NICHD NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- S10 MH124733/MH/NIMH NIH HHS/United States
- R00 HL143036/HL/NHLBI NIH HHS/United States
- R21 NS121642/NS/NINDS NIH HHS/United States
- K99 HL143036/HL/NHLBI NIH HHS/United States
- R01 NS122904/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

