IL-17A-producing γδT cells promote muscle regeneration in a microbiota-dependent manner
- PMID: 35380608
- PMCID: PMC8989184
- DOI: 10.1084/jem.20211504
IL-17A-producing γδT cells promote muscle regeneration in a microbiota-dependent manner
Abstract
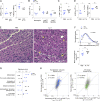
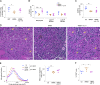
Subsequent to acute injury, skeletal muscle undergoes a stereotypic regenerative process that reestablishes homeostasis. Various types of innate and adaptive immunocytes exert positive or negative influences at specific stages along the course of muscle regeneration. We describe an unanticipated role for γδT cells in promoting healthy tissue recovery after injection of cardiotoxin into murine hindlimb muscle. Within a few days of injury, IL-17A-producing γδT cells displaying primarily Vγ6+ antigen receptors accumulated at the wound site. Punctual ablation experiments showed that these cells boosted early inflammatory events, notably recruitment of neutrophils; fostered the proliferation of muscle stem and progenitor cells; and thereby promoted tissue regeneration. Supplementation of mice harboring low numbers of IL-17A+ γδT cells with recombinant IL-17A largely reversed their inflammatory and reparative defects. Unexpectedly, the accumulation and influences of γδT cells in this experimental context were microbiota dependent, unveiling an orthogonal perspective on the treatment of skeletal muscle pathologies such as catastrophic wounds, wasting, muscular dystrophies, and myositides.
© 2022 Mann et al.
Conflict of interest statement
Disclosures: The authors declare no competing financial interests.
Figures









References
-
- Aguilar, C.A., Shcherbina A., Ricke D.O., Pop R., Carrigan C.T., Gifford C.A., Urso M.L., Kottke M.A., and Meissner A.. 2015. In vivo monitoring of transcriptional dynamics after lower-limb muscle injury enables quantitative classification of healing. Sci. Rep. 5:13885. 10.1038/srep13885 - DOI - PMC - PubMed
-
- Akitsu, A., Ishigame H., Kakuta S., Chung S.H., Ikeda S., Shimizu K., Kubo S., Liu Y., Umemura M., Matsuzaki G., et al. 2015. IL-1 receptor antagonist-deficient mice develop autoimmune arthritis due to intrinsic activation of IL-17-producing CCR2+Vγ6+γδ T cells. Nat. Commun. 6:7464. 10.1038/ncomms8464 - DOI - PMC - PubMed
-
- Alves de Lima, K., Rustenhoven J., Da Mesquita S., Wall M., Salvador A.F., Smirnov I., Cebinelli G.M., Mamuladze T., Baker W., Papadopoulos Z., et al. 2020. Meningeal γδ T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat. Immunol. 21:1421–1429. 10.1038/s41590-020s4150776-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

